4OCV
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JBV
 
 | |
6XN9
 
 | | Solution NMR structure of recifin, a cysteine-rich tyrosyl-DNA Phosphodiesterase I modulatory peptide from the marine sponge Axinella sp. | | Descriptor: | Recifin modulatory peptide | | Authors: | Schroeder, C.I, Rosengren, K.J, O'Keefe, B.R. | | Deposit date: | 2020-07-02 | | Release date: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Recifin A, Initial Example of the Tyr-Lock Peptide Structural Family, Is a Selective Allosteric Inhibitor of Tyrosyl-DNA Phosphodiesterase I.
J.Am.Chem.Soc., 142, 2020
|
|
4JYZ
 
 | |
4KIO
 
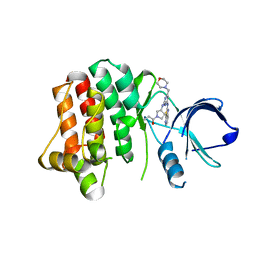 | | Kinase domain mutant of human Itk in complex with a covalently-binding inhibitor | | Descriptor: | 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]prop-2-en-1-one, 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2013-05-02 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of novel irreversible inhibitors of interleukin (IL)-2-inducible tyrosine kinase (Itk) by targeting cysteine 442 in the ATP pocket.
J.Biol.Chem., 288, 2013
|
|
1IJH
 
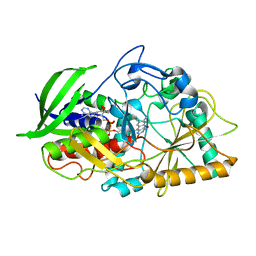 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES ASN485LEU MUTANT | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2001-04-26 | | Release date: | 2001-12-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The presence of a hydrogen bond between asparagine 485 and the pi system of FAD modulates the redox potential in the reaction catalyzed by cholesterol oxidase.
Biochemistry, 40, 2001
|
|
1WND
 
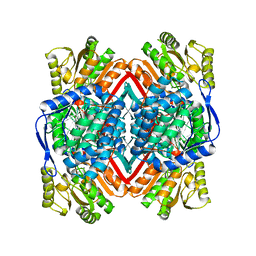 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase as determined by kinetics and crystal structure | | Descriptor: | CALCIUM ION, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
6AGG
 
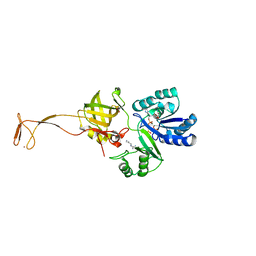 | |
4MAN
 
 | | Bcl_2-Navitoclax Analog (with Indole) Complex | | Descriptor: | 4-[4-({4'-chloro-3-[2-(dimethylamino)ethoxy]biphenyl-2-yl}methyl)piperazin-1-yl]-2-(1H-indol-5-yloxy)-N-({3-nitro-4-[(tetrahydro-2H-pyran-4-ylmethyl)amino]phenyl}sulfonyl)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-08-16 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
1WNB
 
 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase (complexed with nadh and betaine aldehyde) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETAINE ALDEHYDE, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
4PHL
 
 | | TbrPDEB1-inhibitor complex | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, Class 1 phosphodiesterase PDEB1, ETHANOL, ... | | Authors: | Choy, M.S, Bland, N, Peti, W, Page, R. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TbrPDEB1-inhibitor complex
To Be Published
|
|
6S1S
 
 | | Crystal structure of AmpC from Pseudomonas aeruginosa in complex with [3-(2-carboxyvinyl)phenyl]boronic acid] inhibitor | | Descriptor: | (~{E})-3-[3-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Kekez, I, Vicario, M, Bellio, P, Tosoni, E, Celenza, G, Blazquez, J, Tondi, D, Cendron, L. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phenylboronic Acids Probing Molecular Recognition against Class A and Class C beta-lactamases.
Antibiotics, 8, 2019
|
|
5IF0
 
 | |
4LK1
 
 | | Crystal Structure Analysis of the E.coli holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8369 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2Z3S
 
 | | NMR structure of AgTx2-MTX | | Descriptor: | AgTx2-MTX | | Authors: | Pimentel, C, M'Barrek, S, Visan, V, Grissmer, S, Sabatier, J.M, Darbon, H, Fajloun, Z. | | Deposit date: | 2007-06-06 | | Release date: | 2008-04-22 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins.
Protein Sci., 17, 2008
|
|
4Q7Z
 
 | | Neutrophil serine protease 4 (PRSS57) with phe-phe-arg-chloromethylketone (FFR-cmk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
1IGJ
 
 | |
5IDL
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
