3GSB
 
 | |
2MYO
 
 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MYOTROPHIN | | Authors: | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
3VHB
 
 | | IMIDAZOLE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | Descriptor: | IMIDAZOLE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | Deposit date: | 1999-03-17 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
1CML
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1BQQ
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP--TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, MEMBRANE-TYPE MATRIX METALLOPROTEINASE, METALLOPROTEINASE INHIBITOR 2, ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
43CA
 
 | |
1QEY
 
 | | NMR Structure Determination of the Tetramerization Domain of the MNT Repressor: An Asymmetric A-Helical Assembly in Slow Exchange | | Descriptor: | PROTEIN (REGULATORY PROTEIN MNT) | | Authors: | Nooren, I.M.A, George, A.V.E, Kaptein, R, Sauer, R.T, Boelens, R. | | Deposit date: | 1999-04-03 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils.
Nat.Struct.Biol., 6, 1999
|
|
43C9
 
 | |
1CGZ
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH RESVERATROL | | Descriptor: | PROTEIN (CHALCONE SYNTHASE), RESVERATROL | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-26 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1QDL
 
 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CJB
 
 | | MALARIAL PURINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, MAGNESIUM ION, PROTEIN (HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE), ... | | Authors: | Shi, W, Li, C.M, Tyler, P.C, Furneaux, R.H, Cahill, S.M, Girvin, M.E, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-04-08 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor.
Biochemistry, 38, 1999
|
|
1QS0
 
 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | Authors: | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-18 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
1CHW
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH HEXANOYL-COA | | Descriptor: | HEXANOYL-COENZYME A, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1MUY
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI | | Descriptor: | ADENINE GLYCOSYLASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
2U2F
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
1U2F
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
1C46
 
 | |
1C43
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (HUMAN LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C45
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1BQ5
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS GIFU 1051 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Inoue, T, Gotowda, M, Deligeer, Suzuki, S, Kataoka, K, Yamaguchi, K, Watanabe, H, Goho, M, Yasushi, K.A.I. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: comparison of blue and green nitrite reductases.
J.Biochem.(Tokyo), 124, 1998
|
|
1CQV
 
 | |
1GWZ
 
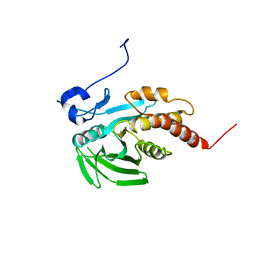 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
1A6B
 
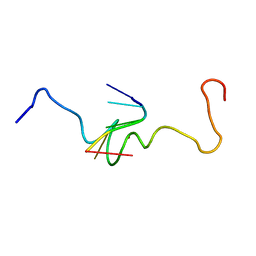 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
1CQ5
 
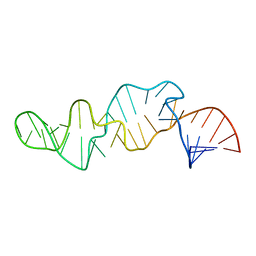 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1CQO
 
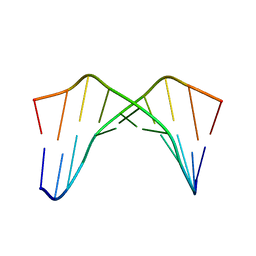 | |
