1L5X
 
 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, Survival protein E | | Authors: | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | Deposit date: | 2002-03-08 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
6GU3
 
 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
1A2W
 
 | | CRYSTAL STRUCTURE OF A 3D DOMAIN-SWAPPED DIMER OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Hart, P.J, Schlunegger, M.P, Eisenberg, D.S. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1LE8
 
 | | Crystal Structure of the MATa1/MATalpha2-3A Heterodimer Bound to DNA Complex | | Descriptor: | 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*CP*AP*TP*CP*A)-3', 5'-D(*TP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*AP*CP*AP*TP*G)-3', MATING-TYPE PROTEIN A-1, ... | | Authors: | Ke, A, Mathias, J.R, Vershon, A.K, Wolberger, C. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Characterization of the DNA Binding Properties of a Triple Alanine Mutant of MATalpha2
Structure, 10, 2002
|
|
2H1W
 
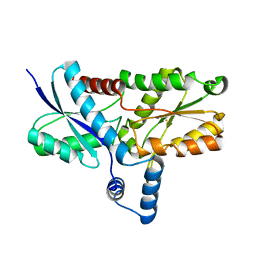 | | Crystal structure of the His183Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | FE (II) ION, Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
1KHO
 
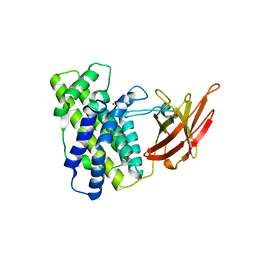 | | Crystal Structure Analysis of Clostridium perfringens alpha-Toxin Isolated from Avian Strain SWCP | | Descriptor: | ZINC ION, alpha-toxin | | Authors: | Justin, N, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The first strain of Clostridium perfringens isolated from an avian source has an alpha-toxin with divergent structural and kinetic properties.
Biochemistry, 41, 2002
|
|
2H4J
 
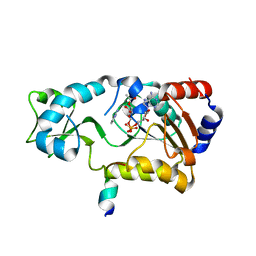 | | Sir2-deacetylated peptide (from enzymatic turnover in crystal) | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, NAD-dependent deacetylase, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2GN4
 
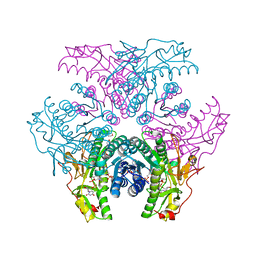 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADPH and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
6GU2
 
 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUB
 
 | | CDK2/CyclinA in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
2H1S
 
 | |
2H2D
 
 | |
2H2F
 
 | |
1KGD
 
 | | Crystal Structure of the Guanylate Kinase-like Domain of Human CASK | | Descriptor: | FORMIC ACID, PERIPHERAL PLASMA MEMBRANE CASK | | Authors: | Li, Y, Spangenberg, O, Paarmann, I, Konrad, M, Lavie, A. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Structural basis for nucleotide-dependent regulation of membrane-associated guanylate kinase-like domains.
J.Biol.Chem., 277, 2002
|
|
1KGP
 
 | | R2F from Corynebacterium Ammoniagenes in its Mn substituted form | | Descriptor: | MANGANESE (II) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
1LQ8
 
 | | Crystal structure of cleaved protein C inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huntington, J.A, Kjellberg, M, Stenflo, J. | | Deposit date: | 2002-05-09 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
6GU7
 
 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
2KU6
 
 | |
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1CMF
 
 | | NMR SOLUTION STRUCTURE OF APO CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Finn, B.E, Evenas, J, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CMG
 
 | | NMR SOLUTION STRUCTURE OF CALCIUM-LOADED CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Evenas, J, Finn, B.E, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
2KU5
 
 | |
2KPF
 
 | | Spatial structure of the dimeric transmembrane domain of glycophorin A in bicelles soluton | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
