4XXD
 
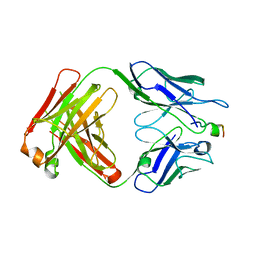 | | Crystal Structure of mid-region amyloid beta capture by solanezumab | | Descriptor: | Amyloid-beta fragment, Fab Heavy Chain, Fab Light Chain | | Authors: | Hermans, S.J, Crespi, G.A.N, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for mid-region amyloid-beta capture by leading Alzheimer's disease immunotherapies.
Sci Rep, 5, 2015
|
|
1HDK
 
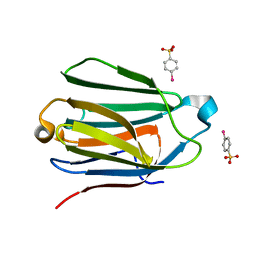 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
7JG1
 
 | | Dimeric Immunoglobin A (dIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7JG2
 
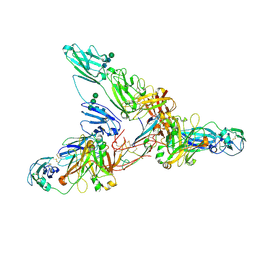 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
2NYT
 
 | | The APOBEC2 Crystal Structure and Functional Implications for AID | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Prochnow, C, Bransteitter, R, Klein, M, Goodman, M, Chen, X. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The APOBEC-2 crystal structure and functional implications for the deaminase AID.
Nature, 445, 2007
|
|
2J88
 
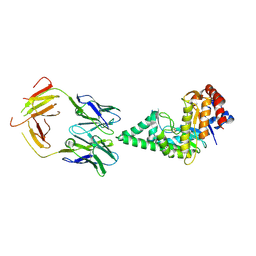 | | Hyaluronidase in complex with a monoclonal IgG Fab fragment | | Descriptor: | FAB, HYALURONONGLUCOSAMINIDASE | | Authors: | Padavattan, S, Schirmer, T, Markovic-Housley, Z. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a B-Cell Epitope of Hyaluronidase, a Major Bee Venom Allergen, from its Crystal Structure in Complex with a Specific Fab.
J.Mol.Biol., 368, 2007
|
|
4TSA
 
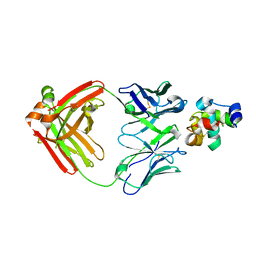 | | Structure of a lysozyme FAb complex | | Descriptor: | FAb Heavy Chain, FAb Light Chain, Lysozyme C | | Authors: | Wensley, B. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a lysozyme FAb complex
To Be Published
|
|
2ZB5
 
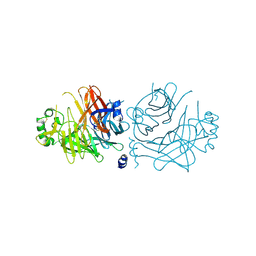 | | Crystal structure of the measles virus hemagglutinin (complex-sugar-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4HBC
 
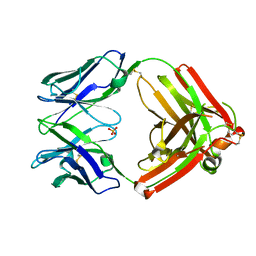 | |
4KHT
 
 | | Triple helix bundle of GP41 complexed with fab 8066 | | Descriptor: | 8066 heavy chain, 8066 light chain, Gp41 helix | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2013-05-01 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41.
Plos One, 8, 2013
|
|
2ZB6
 
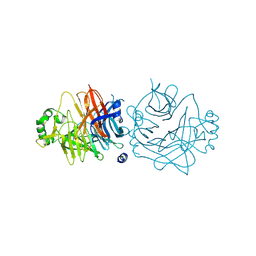 | | Crystal structure of the measles virus hemagglutinin (oligo-sugar type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3DEL
 
 | |
7AQX
 
 | |
7AQY
 
 | |
7AR0
 
 | |
7AQZ
 
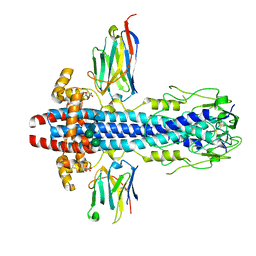 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
7A65
 
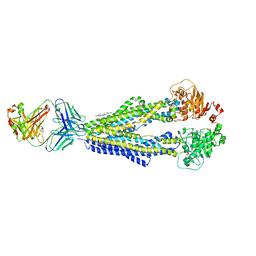 | |
7A6E
 
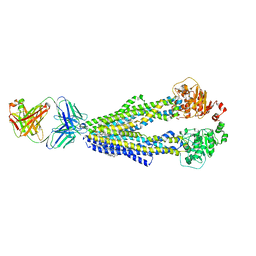 | |
7A25
 
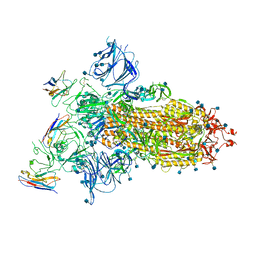 | |
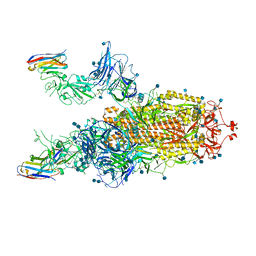
7A69
 
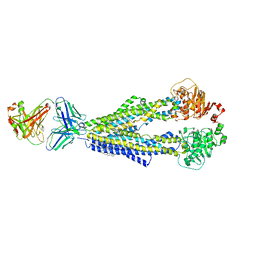 | |
7A6F
 
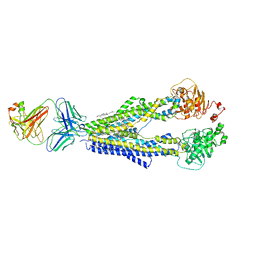 | |
7A6C
 
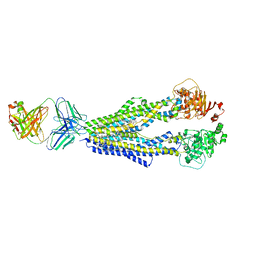 | |
7A29
 
 | |
5NQY
 
 | | Structure of a fHbp(V1.4):PorA(P1.16) chimera. Fusion at fHbp position 309. | | Descriptor: | Factor H binding protein variant B16_001,Major outer membrane protein P.IA,Factor H binding protein variant B16_001 | | Authors: | Johnson, S, Hollingshead, S, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
6DWZ
 
 | | Hermes transposase deletion dimer complex with (C/G) DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
