5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5OYL
 
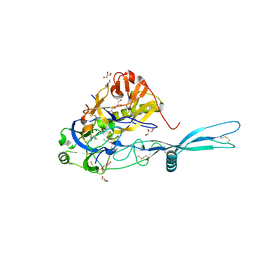 | | VSV G CR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Albertini, A.A, Belot, L, Legrand, P, Gaudin, Y. | | Deposit date: | 2017-09-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein.
Nat Commun, 9, 2018
|
|
3RA8
 
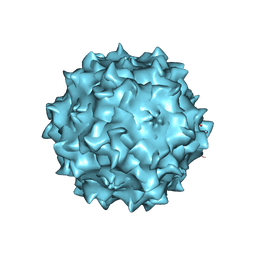 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | ADENOSINE MONOPHOSPHATE, Capsid protein | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3RA2
 
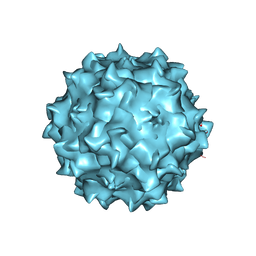 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | Capsid protein | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3RA9
 
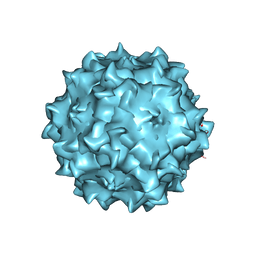 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*A)-3') | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3RAA
 
 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | ADENOSINE MONOPHOSPHATE, Capsid protein | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3RA4
 
 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | Capsid protein, DNA (5'-D(*CP*A)-3') | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3PMK
 
 | | Crystal structure of the Vesicular Stomatitis Virus RNA free nucleoprotein/phosphoprotein complex | | Descriptor: | Nucleocapsid protein, Phosphoprotein | | Authors: | Leyrat, C, Yabukarski, F, Tarbouriech, N, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus N0-P Complex
Plos Pathog., 7, 2011
|
|
6DEJ
 
 | |
5DTL
 
 | |
6WR2
 
 | |
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4R6B
 
 | | Rational Design of Enhanced Photoresistance in a Photoswitchable Fluorescent Protein | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Duan, C, Adam, V, Byrdin, M, Bourgeois, D. | | Deposit date: | 2014-08-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of enhanced photoresistance in a photoswitchable fluorescent protein.
Methods Appl Fluoresc, 3, 2015
|
|
7BYM
 
 | | Cryo-EM structure of human KCNQ4 with retigabine | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYL
 
 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYN
 
 | | Cryo-EM structure of human KCNQ4 with linopirdine | | Descriptor: | 1-phenyl-3,3-bis(pyridin-4-ylmethyl)indol-2-one, Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
6GOY
 
 | | Structure of mEos4b in the green fluorescent state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
2ARL
 
 | | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding | | Descriptor: | ACETIC ACID, CHLORIDE ION, GFP-like non-fluorescent chromoprotein, ... | | Authors: | Wilmann, P.G, Battad, J, Beddoe, T, Olsen, S, Smith, S.C, Dove, S, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2005-08-19 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding
Photochem.Photobiol., 82, 2006
|
|
7FHN
 
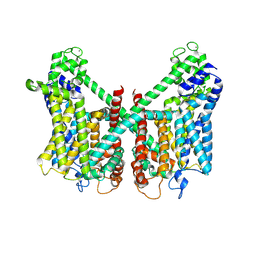 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7X5V
 
 | |
6L26
 
 | | Neutron crystal structure of the mutant green fluorescent protein (EGFP) | | Descriptor: | Green fluorescent protein, trideuteriooxidanium | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.444 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
8UB6
 
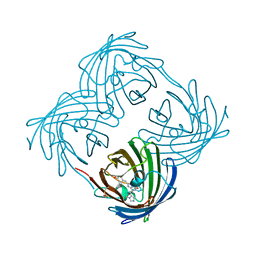 | |
6W22
 
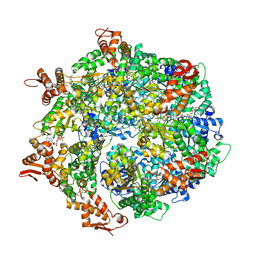 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
6W21
 
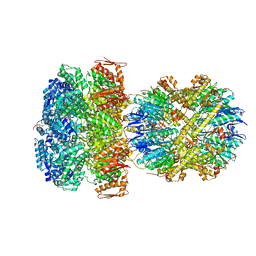 | | ClpAP Engaged2 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
