7O1M
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1J
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7LDU
 
 | |
7LDV
 
 | |
7LDT
 
 | |
7LDW
 
 | |
7LDC
 
 | |
7LD2
 
 | |
7LCL
 
 | |
7LCA
 
 | |
7LBZ
 
 | |
7CW4
 
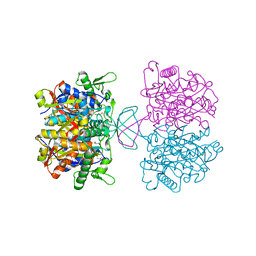 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CW5
 
 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
6L2G
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA-acetyltransferase, putative | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
6PCB
 
 | |
6PCC
 
 | |
6PCD
 
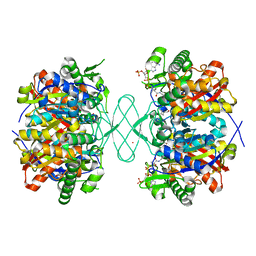 | |
6HSJ
 
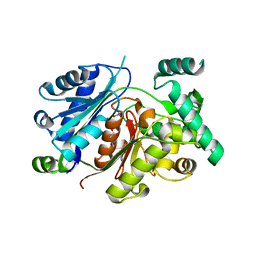 | | Crystal structure of the zebrafish peroxisomal SCP2-thiolase (type-1) in complex with CoA | | Descriptor: | ACETATE ION, COENZYME A, GLYCEROL, ... | | Authors: | Wierenga, R.K, Kiema, T.R, Thapa, C.J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The peroxisomal zebrafish SCP2-thiolase (type-1) is a weak transient dimer as revealed by crystal structures and native mass spectrometry.
Biochem. J., 476, 2019
|
|
6HSP
 
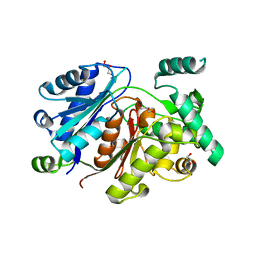 | | Crystal structure of the zebrafish peroxisomal SCP2-thiolase (type-1) in complex with CoA and octanoyl-CoA | | Descriptor: | COENZYME A, GLYCEROL, OCTANOYL-COENZYME A, ... | | Authors: | Wierenga, R.K, Kiema, T.R, Thapa, C.J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The peroxisomal zebrafish SCP2-thiolase (type-1) is a weak transient dimer as revealed by crystal structures and native mass spectrometry.
Biochem. J., 476, 2019
|
|
6HRV
 
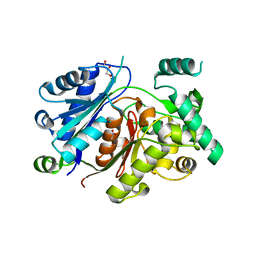 | |
6DV2
 
 | | Crystal Structure of Human Mitochondrial Trifunctional Protein | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Fu, Z, Xia, C, Battaile, K.P, Kim, J.P. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of human mitochondrial trifunctional protein, a fatty acid beta-oxidation metabolon.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZRV
 
 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZQZ
 
 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
