7X9H
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 9 hr using 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WWA
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 2.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW9
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1.5 hr in 20 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW8
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 5 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW7
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW6
 
 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 20 min in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WW5
 
 | | Crystal structure of MutT-8-oxo-dGTP complex | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TN4
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 3-diphosphoinositol 1,2,4,5-tetrakisphosphate (3-PP-IP4), Mg and Fluoride ion | | Descriptor: | (1R,2S,3R,4R,5S,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2022-01-20 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and catalytic analyses of the InsP 6 kinase activities of higher plant ITPKs.
Faseb J., 36, 2022
|
|
7T7H
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7R0D
 
 | |
7NR6
 
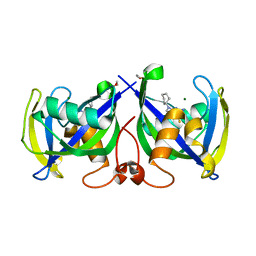 | | Structure of NUDT15 in complex with NSC56456 | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-9-cyclohexyl-3~{H}-purine-6-thione, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling cellular target engagement to drug-induced responses with CeTEAM
To Be Published
|
|
7NNJ
 
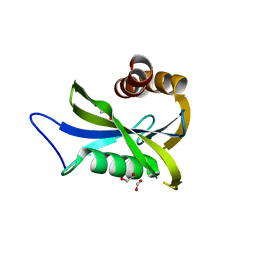 | | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105) | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2, FORMIC ACID, ... | | Authors: | Dubianok, Y, Arruda Bezerra, G, Raux, B, Diaz Saez, L, Riley, A.M, Potter, B.V.L, Huber, K.V.M, von Delft, F. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105)
To Be Published
|
|
7N13
 
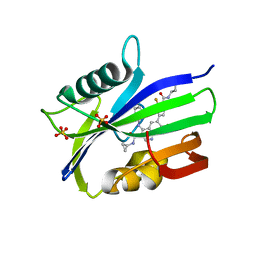 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7N03
 
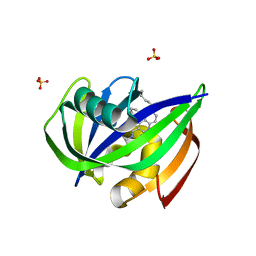 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7ESF
 
 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7E44
 
 | | Crystal structure of NudC complexed with dpCoA | | Descriptor: | DEPHOSPHO COENZYME A, NADH pyrophosphatase, ZINC ION | | Authors: | Zhou, W, Guan, Z.Y, Yin, P, Zhang, D.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into dpCoA-RNA decapping by NudC.
Rna Biol., 18, 2021
|
|
7DNU
 
 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7B7V
 
 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
7B67
 
 | | Structure of NUDT15 V18_V19insGV Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B66
 
 | | Structure of NUDT15 R139H Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B65
 
 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
