6JFP
 
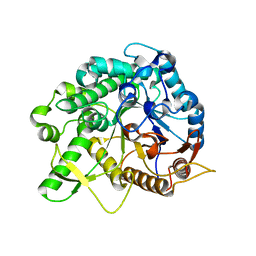 | | Crystal structure of the beta-glucosidase Bgl15 | | Descriptor: | beta-D-glucopyranose, beta-glucosidase 15 | | Authors: | Xie, W, Chen, R. | | Deposit date: | 2019-02-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis.
J.Agric.Food Chem., 68, 2020
|
|
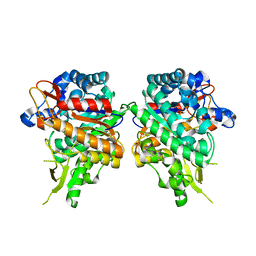
7BBS
 
 | |
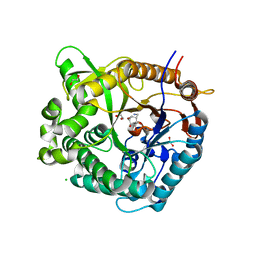
5N6T
 
 | |
6NFJ
 
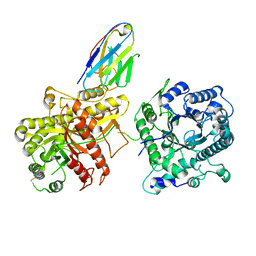 | |
5NAQ
 
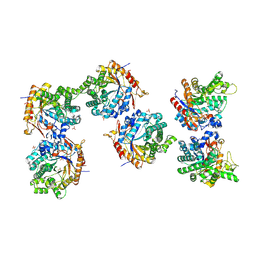 | |
5JBK
 
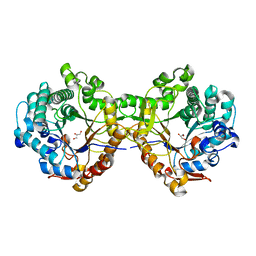 | | Trichoderma harzianum GH1 beta-glucosidase ThBgl1 | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Florindo, R.N, Mutti, H.S, Polikarpov, I, Nascimento, A.S. | | Deposit date: | 2016-04-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum.
N Biotechnol, 40, 2018
|
|
5N6S
 
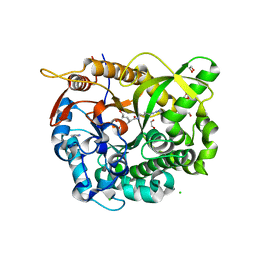 | |
2O9P
 
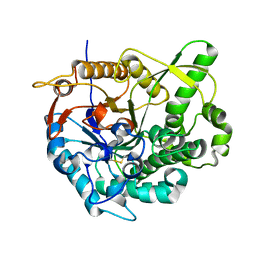 | | beta-glucosidase B from Paenibacillus polymyxa | | Descriptor: | Beta-glucosidase B | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
5NAV
 
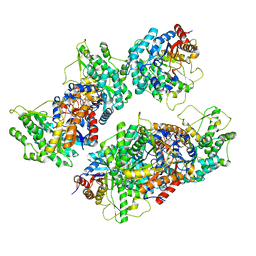 | |
6KHT
 
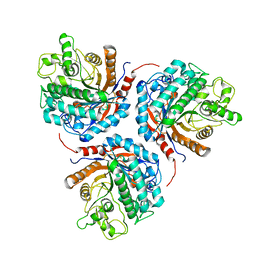 | | Chimeric beta-glucosidase Cel1b-H13 | | Descriptor: | Glycoside hydrolase family 1 | | Authors: | Niu, K.L. | | Deposit date: | 2019-07-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.305 Å) | | Cite: | Chimeric beta-glucosidase Cel1b-H13
To Be Published
|
|
8J3M
 
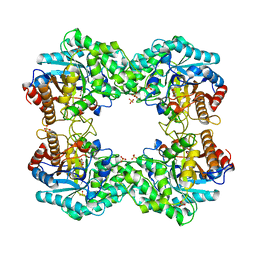 | |
8J5L
 
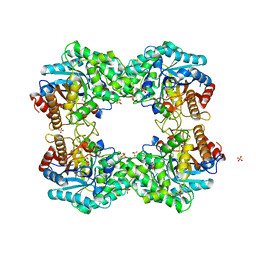 | |
8J5M
 
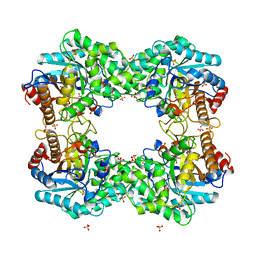 | |
2O9T
 
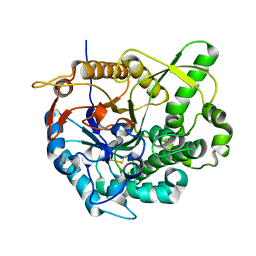 | |
2O9R
 
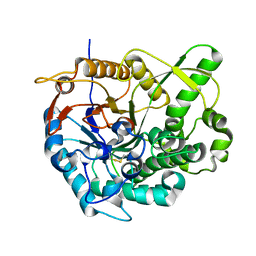 | | beta-glucosidase B complexed with thiocellobiose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
6IER
 
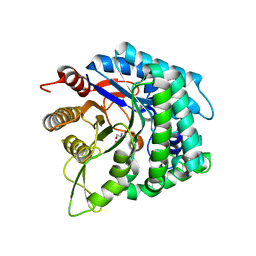 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
5IDI
 
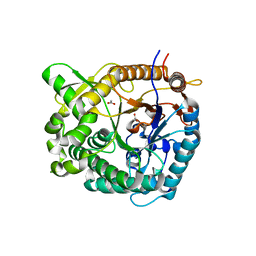 | | Structure of beta glucosidase 1A from Thermotoga neapolitana, mutant E349A | | Descriptor: | 1,4-beta-D-glucan glucohydrolase, ACETATE ION | | Authors: | Kulkarni, T, Nordberg Karlsson, E, Logan, D.T. | | Deposit date: | 2016-02-24 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-glucosidase 1A from Thermotoga neapolitana and comparison of active site mutants for hydrolysis of flavonoid glucosides.
Proteins, 85, 2017
|
|
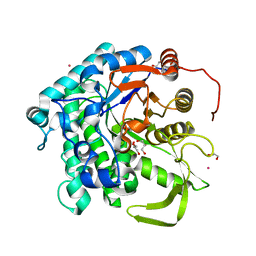
6KDC
 
 | |
2PBG
 
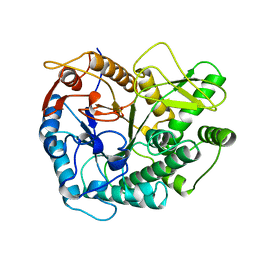 | | 6-PHOSPHO-BETA-D-GALACTOSIDASE FORM-B | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
7D6B
 
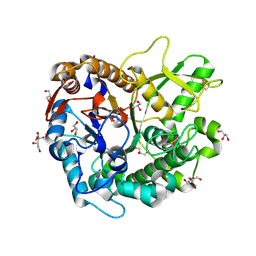 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase with delta-gluconolactone | | Descriptor: | Beta-glucosidase 18, D-glucono-1,5-lactone, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
7D6A
 
 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 18, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
4MDP
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens in complex with glucose | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EAN
 
 | | 1.75A resolution structure of indole bound beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-galactosidase, CHLORIDE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
4EK7
 
 | | High speed X-ray analysis of plant enzymes at room temperature | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
4EAM
 
 | | 1.70A resolution structure of apo beta-glycosidase (W33G) from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Deckert, K, Brunner, L.C, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designing allosteric control into enzymes by chemical rescue of structure.
J.Am.Chem.Soc., 134, 2012
|
|
