5H7V
 
 | |
5H9B
 
 | |
4P4B
 
 | |
4P4D
 
 | |
4P4F
 
 | |
3FEE
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FED
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with a transition state analog of Glu-Glu | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-amino-3-carboxypropyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
4P4I
 
 | |
6KCU
 
 | | Shuguo PWWP domain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Shuguo PWWP domain
To Be Published
|
|
3G7T
 
 | |
6KCO
 
 | | Shuguo PWWP in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*T)-3'), LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shuguo PWWP in complex with ssDNA
To Be Published
|
|
5F84
 
 | |
6I0V
 
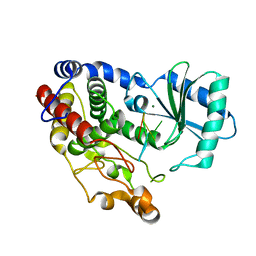 | | Crystal structure of DmTailor in complex with CACAGU RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*AP*CP*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6I0S
 
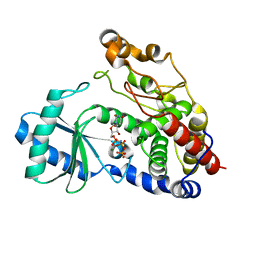 | | Crystal structure of DmTailor in complex with UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
4ZN8
 
 | |
6I0T
 
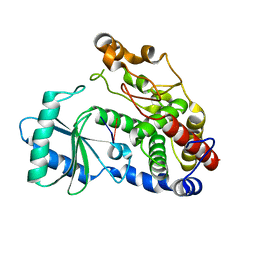 | | Crystal structure of DmTailor in complex with GpU | | Descriptor: | RNA (5'-R(*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
5HLE
 
 | |
4QTR
 
 | | Computational design of co-assembling protein-DNA nanowires | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*TP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*AP*TP*TP*TP*AP*AP*TP*TP*TP*CP*C)-3'), dualENH | | Authors: | Mou, Y, Mayo, S.L. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of co-assembling protein-DNA nanowires.
Nature, 525, 2015
|
|
4QT8
 
 | | Crystal Structure of RON Sema-PSI-IPT1 extracellular domains in complex with MSP beta-chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor-like protein, Macrophage-stimulating protein receptor, ... | | Authors: | Herzberg, O, Chao, K.L. | | Deposit date: | 2014-07-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
J.Biol.Chem., 289, 2014
|
|
4R08
 
 | | Crystal structure of human TLR8 in complex with ssRNA40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3'-O-[(R)-{[(2R,3aR,4R,6R,6aR)-6-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-hydroxy-2-oxidotetrahydrofuro[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]uridine 5'-(dihydrogen phosphate), ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toll-like receptor 8 senses degradation products of single-stranded RNA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6K0O
 
 | | The crystal structure of human CD163-like homolog SRCR8 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M160 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
4EZS
 
 | |
3H3D
 
 | |
4OC3
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CFIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(furan-2-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1T1M
 
 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
