1UQC
 
 | |
1UQE
 
 | |
1UQG
 
 | |
1UQF
 
 | |
1UQA
 
 | |
1UQB
 
 | |
1ERA
 
 | |
4AXP
 
 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
1FRA
 
 | |
5T2P
 
 | | Hepatitis B virus core protein Y132A mutant in complex with sulfamoylbenzamide (SBA_R01) | | Descriptor: | 4-fluoranyl-3-(4-oxidanylpiperidin-1-yl)sulfonyl-~{N}-[3,4,5-tris(fluoranyl)phenyl]benzamide, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
4Q7N
 
 | | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, N,N,N-trimethyl-4-oxobutan-1-aminium | | Authors: | Chaudhary, A, Tyagi, T.K, Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution
To be Published
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
2MJC
 
 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
4NL8
 
 | | PriA Helicase Bound to SSB C-terminal Tail Peptide | | Descriptor: | Primosome assembly protein PriA, Single-stranded DNA-binding protein, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Thurmes, T.M, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.08 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NC9
 
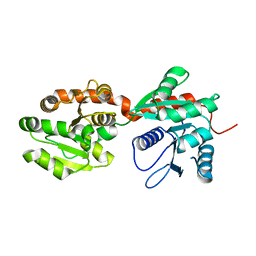 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
4NL4
 
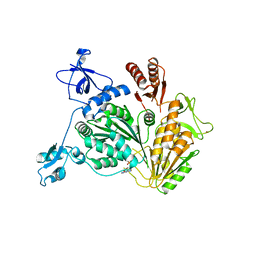 | | PriA Helicase Bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primosome assembly protein PriA, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NET
 
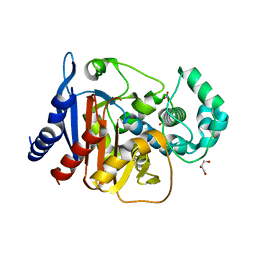 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2OH2
 
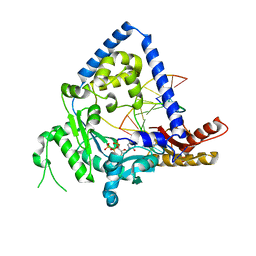 | | Ternary Complex of Human DNA Polymerase | | Descriptor: | 5'-D(*GP*GP*G*GP*GP*AP*AP*GP*GP*AP*CP*CP*C)-3', 5'-D(*TP*T*CP*CP*AP*GP*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', DNA polymerase kappa, ... | | Authors: | Lone, S, Townson, S.A, Uljon, S.N, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2007-01-09 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ternary complex of the catalytic core of human DNA polymerase Kappa with DNA and dTT
To be Published
|
|
4PP4
 
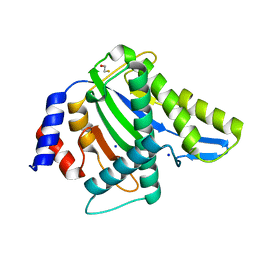 | |
4O8G
 
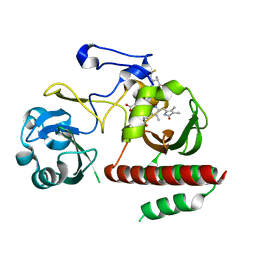 | | Structure of Infrared Fluorescent Protein 1.4 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Bhattacharya, S, Forest, K.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
4O90
 
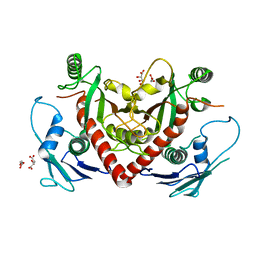 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution | | Descriptor: | Chorismate synthase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Chaudhary, A, Singh, N, Shukla, P.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-31 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution
To be Published
|
|
2WTZ
 
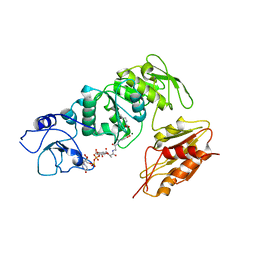 | | MurE ligase of Mycobacterium Tuberculosis | | Descriptor: | MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE-D-GLUTAMATE | | Authors: | Basavannacharya, C, Robertson, G, Munshi, T, Keep, N.H, Bhakta, S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP-Dependent Mure Ligase in Mycobacterium Tuberculosis: Biochemical and Structural Characterisation.
Tuberculosis(Edinb.), 90, 2010
|
|
2WAL
 
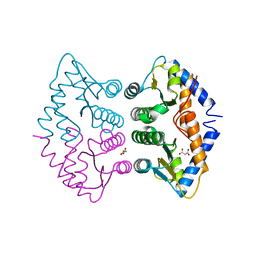 | | Crystal Structure of human GADD45gamma | | Descriptor: | GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE PROTEIN GADD45 GAMMA, MALONIC ACID | | Authors: | Bhattacharya, S, Mueller, J.J, Roske, Y, Turnbull, A.P, Quedenau, C, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Gadd45Gamma
To be Published
|
|
4N9W
 
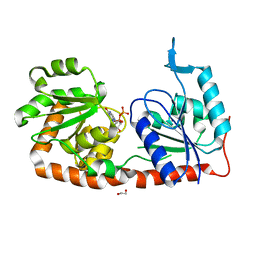 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
2CQ0
 
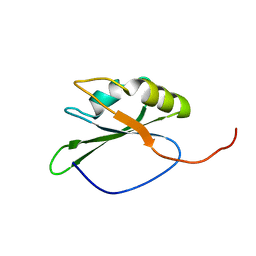 | | solution structure of RNA binding domain in eukaryotic translation initiation factor 3 subunit 4 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit 4 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RNA binding domain in eukaryotic translation initiation factor 3 subunit 4
To be Published
|
|
