2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2VVI
 
 | | IrisFP fluorescent protein in its green form, trans conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VVH
 
 | | IrisFP fluorescent protein in its green form, cis conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VZX
 
 | | Structural and spectroscopic characterization of photoconverting fluorescent protein Dendra2 | | Descriptor: | GLYCEROL, Green fluorescent protein, TETRAETHYLENE GLYCOL | | Authors: | Adam, V, Nienhaus, K, Bourgeois, D, Nienhaus, G.U. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Enhanced Photoconversion Yield in Green Fluorescent Protein-Like Protein Dendra2.
Biochemistry, 48, 2009
|
|
1S6Z
 
 | | Enhanced Green Fluorescent Protein Containing the Y66L Substitution | | Descriptor: | CHLORIDE ION, green fluorescent protein | | Authors: | Rosenow, M.A, Huffman, H.A, Phail, M.E, Wachter, R.M. | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Y66L Variant of Green Fluorescent Protein Supports a Cyclization-Oxidation-Dehydration Mechanism for Chromophore Maturation(,).
Biochemistry, 43, 2004
|
|
8OVN
 
 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OVO
 
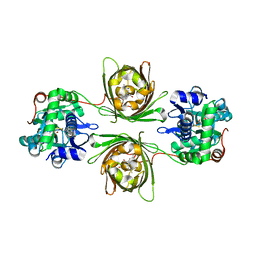 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVP
 
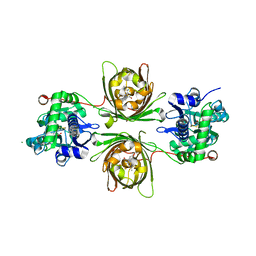 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
5WUA
 
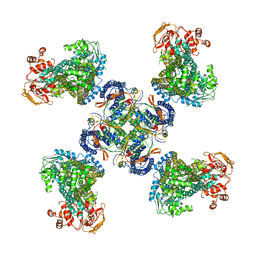 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
8FED
 
 | | Structure of Mce1-LucB complex from Mycobacterium smegmatis (Map1) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEF
 
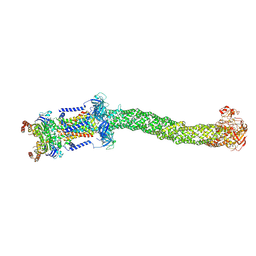 | | Structure of Mce1 transporter from Mycobacterium smegmatis (Map0) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEE
 
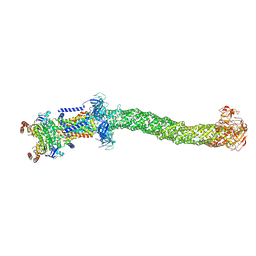 | | Structure of Mce1 transporter from Mycobacterium smegmatis in the absence of LucB (Map2) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
7UGT
 
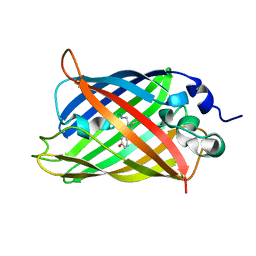 | |
7UGR
 
 | | Crystal structure of hyperfolder YFP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyperfolder yellow fluorescent protein, ... | | Authors: | Campbell, B.C, Liu, C.F, Petsko, G.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chemically stable fluorescent proteins for advanced microscopy.
Nat.Methods, 19, 2022
|
|
6VAL
 
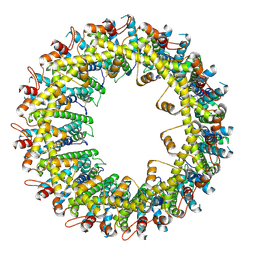 | |
6VAM
 
 | |
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
7PHR
 
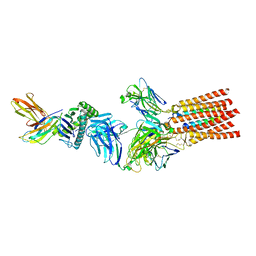 | | Structure of a fully assembled T-cell receptor engaging a tumor-associated peptide-MHC I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Susac, L, Thomas, C, Tampe, R. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC.
Cell, 185, 2022
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXD
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
6QUG
 
 | | GHK tagged MBP-Nup98(1-29) | | Descriptor: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4JF9
 
 | |
4JEO
 
 | |
