1FM2
 
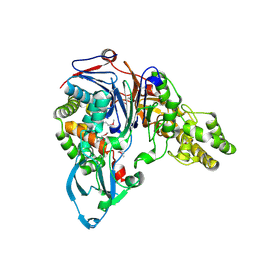 | | THE 2 ANGSTROM CRYSTAL STRUCTURE OF CEPHALOSPORIN ACYLASE | | Descriptor: | GLUTARYL 7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Kim, Y, Yoon, K.H, Khang, Y, Turley, S, Hol, W.G.J. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of cephalosporin acylase.
Structure Fold.Des., 8, 2000
|
|
1FM4
 
 | | CRYSTAL STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 1L | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, MAJOR POLLEN ALLERGEN BET V 1-L | | Authors: | Markovic-Housley, Z, Degano, M, Lamba, D, von Roepenack-Lahaye, E, Clemens, S, Susani, M, Ferreira, F, Scheiner, O, Breiteneder, H. | | Deposit date: | 2000-08-16 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet
v 1 and its Likely Biological Function as a Plant Steroid Carrier
J.Mol.Biol., 325, 2003
|
|
1FM5
 
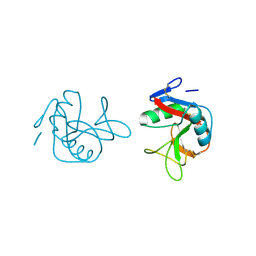 | |
1FM6
 
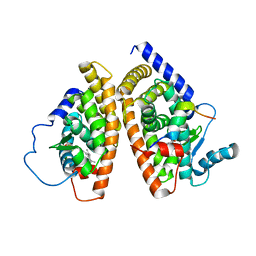 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND ROSIGLITAZONE AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
1FM7
 
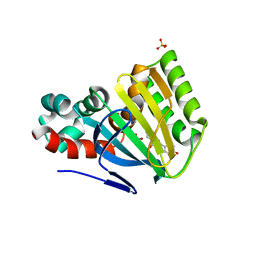 | | CHALCONE ISOMERASE COMPLEXED WITH 5-DEOXYFLAVANONE | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE-FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Noel, J.P. | | Deposit date: | 2000-08-16 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences.
J.Biol.Chem., 277, 2002
|
|
1FM8
 
 | | CHALCONE ISOMERASE COMPLEXED WITH 5,4'-DIDEOXYFLAVANONE | | Descriptor: | 7-HYDROXY-2-PHENYL-CHROMAN-4-ONE, CHALCONE-FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Noel, J.P. | | Deposit date: | 2000-08-16 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences.
J.Biol.Chem., 277, 2002
|
|
1FM9
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA, ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
1FMA
 
 | | MOLYBDOPTERIN SYNTHASE (MOAD/MOAE) | | Descriptor: | CHLORIDE ION, MOLYBDOPTERIN CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Rudolph, M.J, Wuebbens, M.M, Rajagolpalan, K.V, Schindelin, H. | | Deposit date: | 2000-08-16 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation.
Nat.Struct.Biol., 8, 2001
|
|
1FMB
 
 | | EIAV PROTEASE COMPLEXED WITH THE INHIBITOR HBY-793 | | Descriptor: | EIAV PROTEASE, [2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYL-VALINYL]-PHENYLALANINOL | | Authors: | Wlodawer, A, Gustchina, A, Zdanov, A, Kervinen, J. | | Deposit date: | 1996-02-27 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of equine infectious anemia virus proteinase complexed with an inhibitor.
Protein Sci., 5, 1996
|
|
1FMC
 
 | | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEX WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
1FMD
 
 | | THE STRUCTURE AND ANTIGENICITY OF A TYPE C FOOT-AND-MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Lea, S, Fry, E, Stuart, D. | | Deposit date: | 1994-02-10 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure and antigenicity of a type C foot-and-mouth disease virus.
Structure, 2, 1994
|
|
1FME
 
 | |
1FMF
 
 | | REFINED SOLUTION STRUCTURE OF THE (13C,15N-LABELED) B12-BINDING SUBUNIT OF GLUTAMATE MUTASE FROM CLOSTRIDIUM TETANOMORPHUM | | Descriptor: | METHYLASPARTATE MUTASE S CHAIN | | Authors: | Hoffmann, B, Konrat, R, Tollinger, M, Huhta, M, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 2000-08-17 | | Release date: | 2002-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A protein pre-organized to trap the nucleotide moiety of coenzyme B(12): refined solution structure of the B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum.
Chembiochem, 2, 2001
|
|
1FMG
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.04% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-17 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FMH
 
 | |
1FMI
 
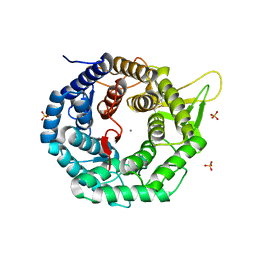 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE | | Descriptor: | CALCIUM ION, ENDOPLASMIC RETICULUM ALPHA-MANNOSIDASE I, SULFATE ION | | Authors: | Vallee, F, Karaveg, K, Herscovics, A, Moremen, K.W, Howell, P.L. | | Deposit date: | 2000-08-17 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
1FMJ
 
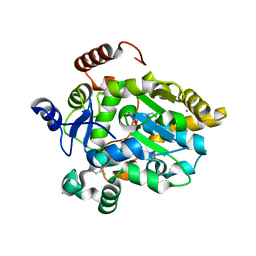 | | CRYSTAL STRUCTURE OF MERCURY DERIVATIVE OF RETINOL DEHYDRATASE IN A COMPLEX WITH RETINOL AND PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Pakhomova, S, Kobayashi, M, Buck, J, Newcomer, M.E. | | Deposit date: | 2000-08-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A helical lid converts a sulfotransferase to a dehydratase.
Nat.Struct.Biol., 8, 2001
|
|
1FMK
 
 | |
1FML
 
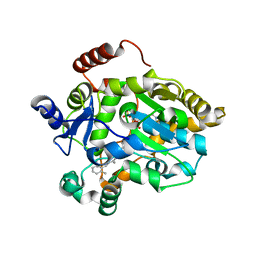 | | CRYSTAL STRUCTURE OF RETINOL DEHYDRATASE IN A COMPLEX WITH RETINOL AND PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, RETINOL, ... | | Authors: | Pakhomova, S, Kobayashi, M, Buck, J, Newcomer, M.E. | | Deposit date: | 2000-08-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A helical lid converts a sulfotransferase to a dehydratase.
Nat.Struct.Biol., 8, 2001
|
|
1FMM
 
 | | SOLUTION STRUCTURE OF NFGF-1 | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Arunkumar, A.I, Srisailam, S, Kumar, T.K.S, Chiu, I.M, Yu, C. | | Deposit date: | 2000-08-18 | | Release date: | 2001-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens.
J.Biol.Chem., 277, 2002
|
|
1FMN
 
 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
1FMO
 
 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
1FMQ
 
 | | Cyclo-butyl-bis-furamidine complexed with dCGCGAATTCGCG | | Descriptor: | 2,5-BIS{[4-(N-CYCLOBUTYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
1FMS
 
 | | Structure of complex between cyclohexyl-bis-furamidine and d(CGCGAATTCGCG) | | Descriptor: | 2,5-BIS{[4-(N-CYCLOHEXYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
1FMT
 
 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA FMET FORMYLTRANSFERASE | | Authors: | Schmitt, E, Mechulam, Y. | | Deposit date: | 1997-10-13 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase.
EMBO J., 15, 1996
|
|
