6BJE
 
 | |
1MQB
 
 | | Crystal Structure of Ephrin A2 (ephA2) Receptor Protein Kinase | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N, Rogers, J, Sang, B.C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Cancer Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2003
|
|
7ES0
 
 | | a rice glycosyltransferase in complex with UDP and REX | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
2K8C
 
 | | Solution structure of PLAA family ubiquitin binding domain (PFUC) trans isomer in complex with ubiquitin | | Descriptor: | Phospholipase A-2-activating protein, Ubiquitin | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2K89
 
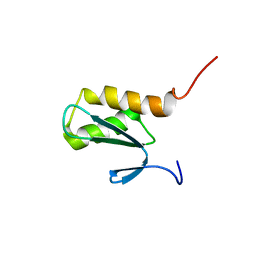 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 cis isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2K8B
 
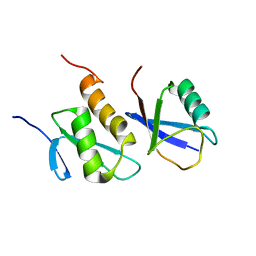 | | Solution structure of PLAA family ubiquitin binding domain (PFUC) cis isomer in complex with ubiquitin | | Descriptor: | Phospholipase A-2-activating protein, Ubiquitin | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
5WSH
 
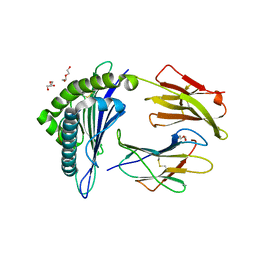 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
4L3E
 
 | |
2K8A
 
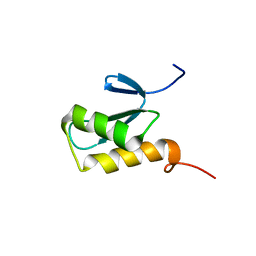 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 trans isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
4L3B
 
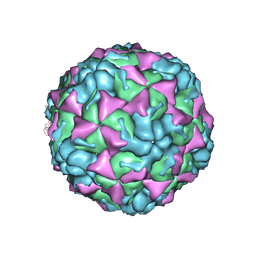 | | X-ray structure of the HRV2 A particle uncoating intermediate | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Vives-Adrian, L, Querol-Audi, J, Garriga, D, Pous, J, Verdaguer, N. | | Deposit date: | 2013-06-05 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1I1Y
 
 | |
1I1F
 
 | | Crystal structure of human class i mhc (hla-a2.1) complexed with beta 2-microglobulin and hiv-rt variant peptide i1y | | Descriptor: | PROTEIN (BETA 2-MICROGLOBULIN), PROTEIN (CLASS I HISTOCOMPATIBILITY ANTIGEN, GOGO-A0201 ALPHA CHAIN), ... | | Authors: | Kirksey, T.J, Pogue-Caley, R.R, Frelinger, J.A, Collins, E.J. | | Deposit date: | 1999-04-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the increased immunogenicity of two HIV-reverse transcriptase peptide variant/class I major histocompatibility complexes.
J.Biol.Chem., 274, 1999
|
|
8XD7
 
 | |
8XDB
 
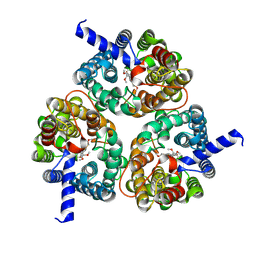 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | (5E)-2-azanylidene-5-[(2,3-dimethoxyphenyl)methylidene]-1,3-thiazolidin-4-one, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDC
 
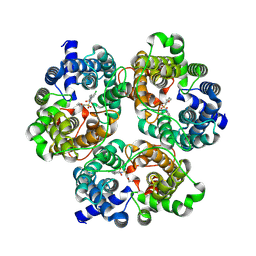 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
3P70
 
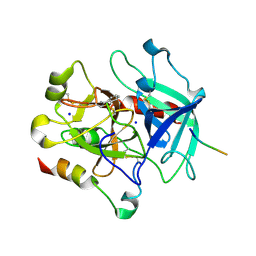 | | Structural basis of thrombin-mediated factor V activation: essential role of the hirudin-like sequence Glu666-Glu672 for processing at the heavy chain-B domain junction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Corral-Rodriguez, M.A, Bock, P.E, Hernandez-Carvajal, E, Gutierrez-Gallego, R, Fuentes-Prior, P. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of thrombin-mediated factor V activation: the Glu666-Glu672 sequence is critical for processing at the heavy chain-B domain junction.
Blood, 117, 2011
|
|
1XRK
 
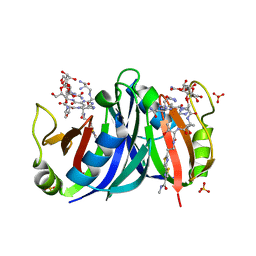 | | Crystal structure of a mutant bleomycin binding protein from Streptoalloteichus hindustanus displaying increased thermostability | | Descriptor: | BLEOMYCIN A2, Bleomycin resistance protein, SULFATE ION | | Authors: | Brouns, S.J.J, Wu, H, Akerboom, J, Turnbull, A.P, de Vos, W.M, Van der Oost, J. | | Deposit date: | 2004-10-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a selectable marker for hyperthermophiles
J.Biol.Chem., 280, 2005
|
|
3TO2
 
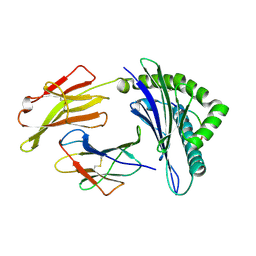 | | Structure of HLA-A*0201 complexed with peptide Md3-C9 derived from a clustering region of restricted cytotoxic T lymphocyte epitope from SARS-CoV M protein | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Md3-C9 peptide derived from Membrane glycoprotein | | Authors: | Liu, J, Qi, J, Gao, F, Yan, J, Gao, G.F. | | Deposit date: | 2011-09-03 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Definition of a Clustering Region of HLA-A2-restricted Cytotoxic T Lymphocyte Epitopes
Sci.Technology Rev., 29, 2011
|
|
4DIF
 
 | | Structure of A1-type ketoreductase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Keatinge-Clay, A.T. | | Deposit date: | 2012-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and mutagenesis of A2-type ketoreductase from modular polyketide synthase reveals insights into stereospecificity
To be Published
|
|
1EWJ
 
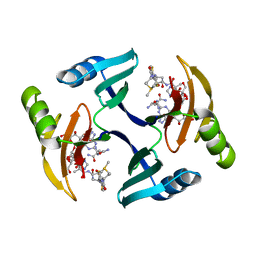 | | CRYSTAL STRUCTURE OF BLEOMYCIN-BINDING PROTEIN COMPLEXED WITH BLEOMYCIN | | Descriptor: | BLEOMYCIN A2, BLEOMYCIN RESISTANCE DETERMINANT | | Authors: | Maruyama, M, Kumagai, T, Matoba, Y, Hata, Y, Sugiyama, M. | | Deposit date: | 2000-04-26 | | Release date: | 2001-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
7KNW
 
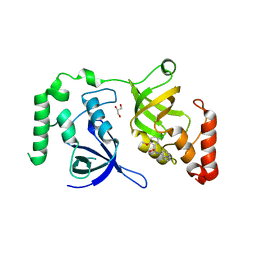 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
1MXK
 
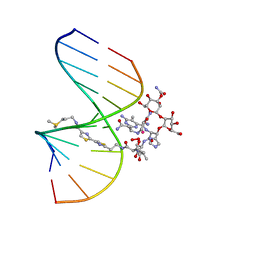 | | NMR Structure of HO2-Co(III)bleomycin A(2) Bound to d(GGAAGCTTCC)(2) | | Descriptor: | 5'-D(*GP*GP*AP*AP*GP*CP*TP*TP*CP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-10-02 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
1MTG
 
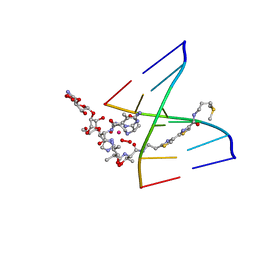 | | NMR Structure of HO2-Co(III)bleomycin A(2) bound to d(GAGCTC)(2) | | Descriptor: | 5'-D(*GP*AP*GP*CP*TP*C)-3', BLEOMYCIN A2, COBALT (III) ION, ... | | Authors: | Zhao, C, Xia, C, Mao, Q, Forsterling, H, DeRose, E, Antholine, W.E, Subczynski, W.K, Petering, D.H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of HO(2)-Co(III)bleomycin A(2) Bound to d(GAGCTC)(2) and d(GGAAGCTTCC)(2): Structure-Reactivity Relationships of Co and Fe Bleomycins
J.Inorg.Biochem., 91, 2002
|
|
1NIQ
 
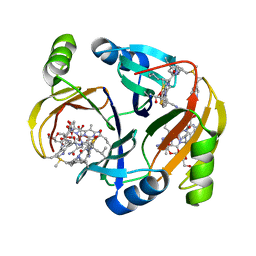 | |
1LKQ
 
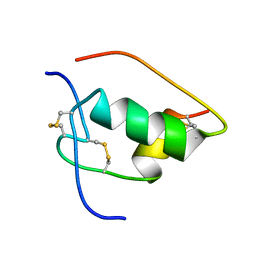 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Weiss, M.A, Hua, Q.X, Chu, Y.C, Jia, W, Philips, N.F, Wang, R.Y, Katsoyannis, P.G. | | Deposit date: | 2002-04-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing
J.Biol.Chem., 277, 2002
|
|
