8R1A
 
 | | Model of the membrane-bound GBP1 oligomer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate binding protein 1, ... | | Authors: | Weismehl, M, Chu, X, Kutsch, M, Lauterjung, P, Herrmann, C, Kudryashev, M, Daumke, O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (26.799999 Å) | | Cite: | Structural insights into the activation mechanism of antimicrobial GBP1.
Embo J., 43, 2024
|
|
8RRR
 
 | | Alpha-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Saibil, H.R, Monistrol, J. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise recruitment of Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibre disassembly
Biorxiv, 2024
|
|
8RQM
 
 | | Alpha-synuclein amyloid fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Saibil, H.R, Monistrol, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise recruitment of Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibre disassembly
Biorxiv, 2024
|
|
7Y1T
 
 | | Complex of integrin alphaV/beta8 and L-TGF-beta1 at a ratio of 1:2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y1R
 
 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y04
 
 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7YAG
 
 | | CryoEM structure of SPCA1a in E1-Ca-AMPPCP state subclass 1 | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
7YAH
 
 | | CryoEM structure of SPCA1a in E1-Ca-AMPPCP state subclass 2 | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
7YAI
 
 | | CryoEM structure of SPCA1a in E1-Ca-AMPPCP state subclass 3 | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, Nanobody head piece of megabody, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
7YAM
 
 | | CryoEM structure of SPCA1a in E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
7YAJ
 
 | | CryoEM structure of SPCA1a in E1-Mn-AMPPCP state subclass 1 | | Descriptor: | Calcium-transporting ATPase type 2C member 1, MANGANESE (II) ION, Nanobody head piece of megabody, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
7XX6
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | Descriptor: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
6ZWO
 
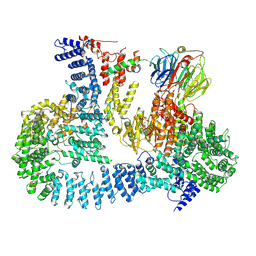 | | cryo-EM structure of human mTOR complex 2, focused on one half | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
6ZWM
 
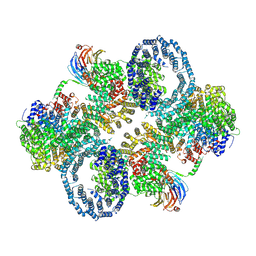 | | cryo-EM structure of human mTOR complex 2, overall refinement | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
3UUL
 
 | |
2DFS
 
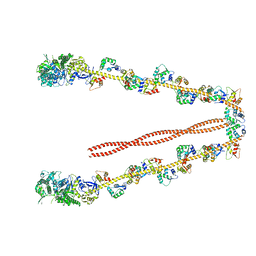 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
1HJ0
 
 | | Thymosin beta9 | | Descriptor: | THYMOSIN BETA9 | | Authors: | Stoll, R, Voelter, W, Holak, T.A. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Conformation of Thymosin Beta9 in Water/Fluoroalcohol Solution Determined by NMR Spectroscopy
Biopolymers, 41, 1997
|
|
8FCA
 
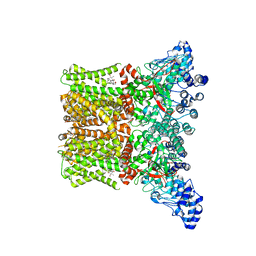 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with 4alpha-Phorbol 12,13-didecanoate | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FC8
 
 | | Cryo-EM structure of the human TRPV4 in complex with GSK1016790A | | Descriptor: | CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
1UJO
 
 | |
4GZM
 
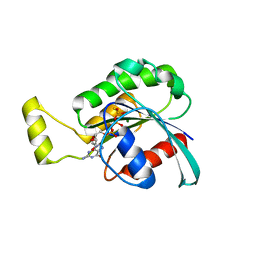 | | Crystal structure of RAC1 F28L mutant | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1UEG
 
 | |
4GZL
 
 | | Crystal structure of RAC1 Q61L mutant | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8E1H
 
 | | Asp1 kinase in complex with ADP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
