1H72
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH HSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HOMOSERINE KINASE, L-HOMOSERINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
3LHA
 
 | |
1EHC
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Jiang, M, Bourret, R, Simon, M, Volz, K. | | Deposit date: | 1996-03-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY.
J.Biol.Chem., 272, 1997
|
|
1H73
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH THREONINE | | Descriptor: | HOMOSERINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THREONINE | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
1HNF
 
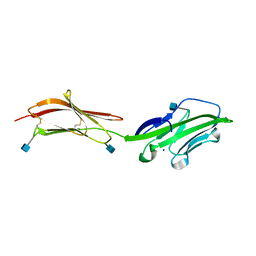 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
5XD1
 
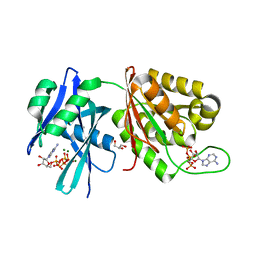 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and magnesium | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD3
 
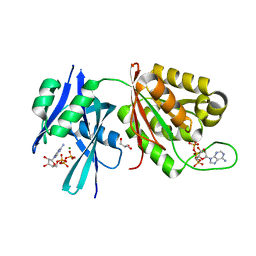 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XD5
 
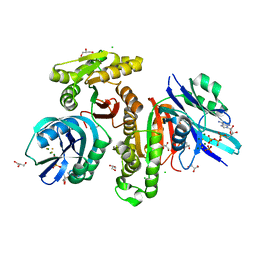 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP, magnesium fluoride and phosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
3QBX
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to 1,6-anhydro-n-actetylmuramic acid | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
2YWQ
 
 | | Crystal structure of Thermus thermophilus Protein Y N-terminal domain | | Descriptor: | Ribosomal subunit interface protein | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Tatsuguchi, A, Saito, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-21 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of Thermus thermophilus Protein Y N-terminal domain
To be Published
|
|
5XD4
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP (II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
2RE9
 
 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
4M4X
 
 | |
4HLY
 
 | |
2MDR
 
 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1FN9
 
 | | CRYSTAL STRUCTURE OF THE REOVIRUS OUTER CAPSID PROTEIN SIGMA 3 | | Descriptor: | OUTER-CAPSID PROTEIN SIGMA 3, ZINC ION | | Authors: | Olland, A.M, Jane-Valbuena, J, Schiff, L.A, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution.
EMBO J., 20, 2001
|
|
4HLX
 
 | |
2N28
 
 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2MS7
 
 | | High-resolution solid-state NMR structure of the helical signal transduction filament MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | He, L, Bardiaux, B, Spehr, J, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2NUF
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.5-Angstrom Resolution | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
3S2I
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
2NUG
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 1.7-Angstrom Resolution | | Descriptor: | 5'-R(P*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*G)-3', 5'-R(P*AP*GP*UP*GP*GP*CP*CP*UP*UP*GP*C)-3', MAGNESIUM ION, ... | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
3S2F
 
 | | Crystal Structure of FurX NADH:Furfural | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHORYLISOPROPANE, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S2G
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol I | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3DUH
 
 | | Structure of Interleukin-23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit beta, Interleukin-23 subunit alpha | | Authors: | Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12.
J.Mol.Biol., 382, 2008
|
|
