2MHG
 
 | |
5PCI
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 39) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4E7I
 
 | | PFV intasome freeze-trapped prior to 3'-processing, Mn-bound form (UI-Mn) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*AP*AP*T)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5301 Å) | | Cite: | 3'-Processing and strand transfer catalysed by retroviral integrase in crystallo.
Embo J., 31, 2012
|
|
3DA2
 
 | | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 13, N-(4-chlorobenzyl)-N-methylbenzene-1,4-disulfonamide, ... | | Authors: | Pilka, E.S, Picaud, S.S, Yue, W.W, King, O.N.F, Bray, J.E, Filippakopoulos, P, Roos, A.K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor.
To be Published
|
|
5PD2
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 58) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDI
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 74) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PE1
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 94) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2RCX
 
 | | AmpC Beta-lactamase in complex with (1R)-1-(2-Thiophen-2-yl-acetylamino)-1-(3-(2-carboxyvinyl)-phenyl) methylboronic acid | | Descriptor: | (1R)-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-1-(3-(2-CARBOXYVINYL)-PHENYL) METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Morandi, F, Morandi, S, Prati, F, Shoichet, B.K. | | Deposit date: | 2007-09-20 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based optimization of cephalothin-analogue boronic acids as beta-lactamase inhibitors
Bioorg.Med.Chem., 16, 2008
|
|
6ZZW
 
 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) and CAS No 912569-62-1 | | Descriptor: | Lectin, SODIUM ION, [3-(2-methylimidazol-1-yl)phenyl]methanamine, ... | | Authors: | Lal, K, Bermeo, R, Imberty, A, Varrot, A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction and Validation of a Druggable Site on Virulence Factor of Drug Resistant Burkholderia cenocepacia*.
Chemistry, 27, 2021
|
|
1PZO
 
 | | TEM-1 Beta-Lactamase in Complex with a Novel, Core-Disrupting, Allosteric Inhibitor | | Descriptor: | Beta-lactamase TEM, N,N-BIS(4-CHLOROBENZYL)-1H-1,2,3,4-TETRAAZOL-5-AMINE | | Authors: | Horn, J.R, Shoichet, B.K. | | Deposit date: | 2003-07-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric inhibition through core disruption.
J.Mol.Biol., 336, 2004
|
|
4Y26
 
 | | Complex of human Galectin-7 and Galbeta1-3(6OSO3)GlcNAc | | Descriptor: | Galectin-7, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
3DFP
 
 | |
3D93
 
 | |
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
2RF2
 
 | | HIV reverse transcriptase in complex with inhibitor 7e (NNRTI) | | Descriptor: | 5-bromo-3-(pyrrolidin-1-ylsulfonyl)-1H-indole-2-carboxamide, Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC 2.7.7.7) (EC 3.1.26.4) (p66 RT) | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2007-09-27 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel indole-3-sulfonamides as potent HIV non-nucleoside reverse transcriptase inhibitors (NNRTIs).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MNV
 
 | | Crystal structure of bicyclic peptide UK729 bound as an acyl-enzyme intermediate to urokinase-type plasminogen activator (uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, ACETATE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5GJR
 
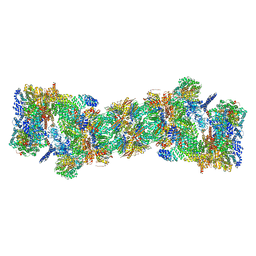 | | An atomic structure of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
2RKU
 
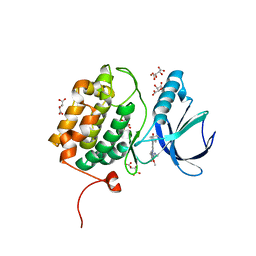 | | Structure of PLK1 in complex with BI2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Ding, Y.-H, Kothe, M, Kohls, D, Low, S. | | Deposit date: | 2007-10-17 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selectivity-determining residues in Plk1.
Chem.Biol.Drug Des., 70, 2007
|
|
7GY7
 
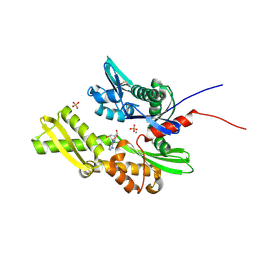 | | Crystal Structure of HSP72 in complex with ligand 10 at 4.56 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GYB
 
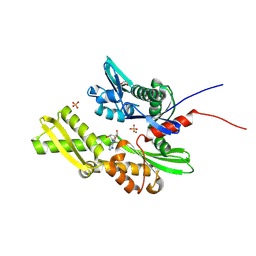 | | Crystal Structure of HSP72 in complex with ligand 10 at 9.12 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
2DA5
 
 | | Solution structure of the second homeobox domain of Zinc fingers and homeoboxes protein 3 (Triple homeobox 1 protein) | | Descriptor: | Zinc fingers and homeoboxes protein 3 | | Authors: | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second homeobox domain of Zinc fingers and homeoboxes protein 3 (Triple homeobox 1 protein)
To be Published
|
|
7DAN
 
 | |
7DG2
 
 | | Nse1-Nse3-Nse4 complex | | Descriptor: | ACETATE ION, GLYCEROL, MAGE domain-containing protein, ... | | Authors: | Cho, Y, Jo, A. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure Basis for Shaping the Nse4 Protein by the Nse1 and Nse3 Dimer within the Smc5/6 Complex.
J.Mol.Biol., 433, 2021
|
|
1PQN
 
 | | dominant negative human hDim1 (hDim1 1-128) | | Descriptor: | Spliceosomal U5 snRNP-specific 15 kDa protein | | Authors: | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
3P9J
 
 | | Aurora A kinase domain with phthalazinone pyrazole inhibitor | | Descriptor: | 4-[(5-methyl-1H-pyrazol-3-yl)amino]-2-phenylphthalazin-1(2H)-one, Serine/threonine-protein kinase 6 | | Authors: | Kairies, N.A, Oliveira, T, Engh, R.A. | | Deposit date: | 2010-10-17 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phthalazinone pyrazoles as potent, selective, and orally bioavailable inhibitors of aurora-a kinase.
J.Med.Chem., 54, 2011
|
|
