7R3J
 
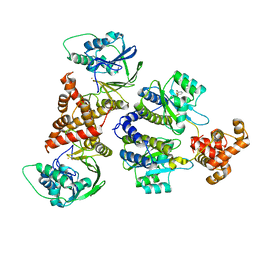 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
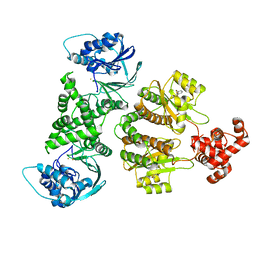 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
5XAZ
 
 | |
5XAY
 
 | |
2FJR
 
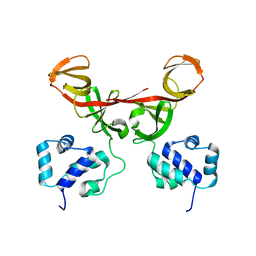 | | Crystal Structure of Bacteriophage 186 | | Descriptor: | Repressor protein CI | | Authors: | Lewis, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of cooperative regulation at an alternate genetic switch
Mol.Cell, 21, 2006
|
|
4ESF
 
 | |
4ESB
 
 | | Crystal structure of PadR-like transcriptional regulator (BC4206) from Bacillus cereus strain ATCC 14579 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Fibriansah, G, Kovacs, A.T, Kuipers, O.P, Thunnissen, A.M.W.H. | | Deposit date: | 2012-04-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Two Transcriptional Regulators from Bacillus cereus Define the Conserved Structural Features of a PadR Subfamily.
Plos One, 7, 2012
|
|
8Q9N
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and MITR deacetylase binding motif mutant L151V. | | Descriptor: | DIMETHYL SULFOXIDE, DNA MADS box, MEF2D protein, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
9FDD
 
 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-05-16 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
9F14
 
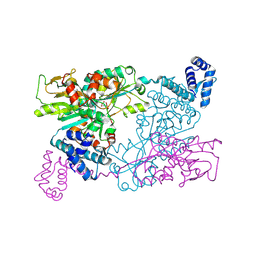 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
2OVM
 
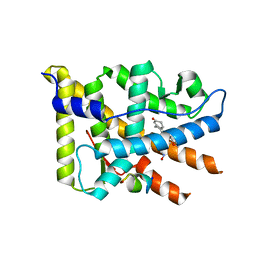 | | Progesterone Receptor with Bound Asoprisnil and a Peptide from the Co-Repressor NCoR | | Descriptor: | 4-[(11BETA,17BETA)-17-METHOXY-17-(METHOXYMETHYL)-3-OXOESTRA-4,9-DIEN-11-YL]BENZALDEHYDE OXIME, NCoR, Progesterone receptor | | Authors: | Madauss, K.P, Deng, S.-J, Short, S.A, Stewart, E.L, Williams, S.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural and in vitro characterization of asoprisnil: a selective progesterone receptor modulator.
Mol.Endocrinol., 21, 2007
|
|
2OVH
 
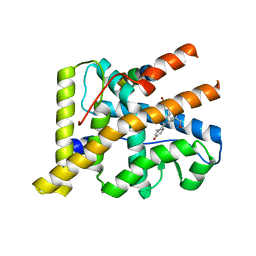 | | Progesterone Receptor with Bound Asoprisnil and a Peptide from the Co-Repressor SMRT | | Descriptor: | 4-[(11BETA,17BETA)-17-METHOXY-17-(METHOXYMETHYL)-3-OXOESTRA-4,9-DIEN-11-YL]BENZALDEHYDE OXIME, Progesterone receptor, SMRT peptide | | Authors: | Madauss, K.P, Deng, S.-J, Short, S.A, Stewart, E.L, Williams, S.P. | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and in vitro characterization of asoprisnil: a selective progesterone receptor modulator.
Mol.Endocrinol., 21, 2007
|
|
1D7V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
6KCR
 
 | |
6ZAB
 
 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (P6422 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Naretto, A, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6ZA7
 
 | | Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
1U9O
 
 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
5J1R
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one at 1.92A resolution | | Descriptor: | 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one, EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5J3L
 
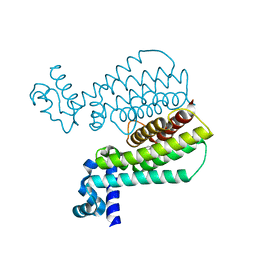 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 1-((2-cyclopentylethyl)sulfonyl)pyrrolidine at 1.66A resolution | | Descriptor: | 1-[(2-cyclopentylethyl)sulfonyl]pyrrolidine, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
8C7U
 
 | |
8C7T
 
 | |
6EL2
 
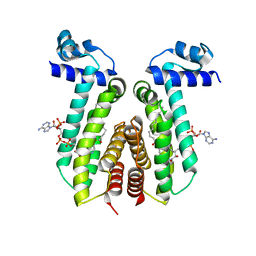 | | SaFadR_lauroyl_CoA complex | | Descriptor: | DODECYL-COA, Transcriptional regulator TetR family | | Authors: | Valegard, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius.
Nat Commun, 10, 2019
|
|
