4Y8F
 
 | |
2JV4
 
 | | Structure Characterisation of PINA WW Domain and Comparison with other Group IV WW Domains, PIN1 and ESS1 | | Descriptor: | Peptidyl-prolyl cis/trans isomerase | | Authors: | Ng, C.A, Kato, Y, Tanokura, M, Brownlee, R.T.C. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of PinA WW domain and a comparison with other Group IV WW domains, Pin1 and Ess1
Biochim.Biophys.Acta, 1784, 2008
|
|
2JZV
 
 | | Solution structure of S. aureus PrsA-PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Seppala, R, Tossavainen, H, Heikkinen, S, Koskela, H, Kontinen, V, Permi, P. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA - Implications for the catalytic mechanism of parvulins.
Bmc Struct.Biol., 9, 2009
|
|
2JFW
 
 | |
2JGQ
 
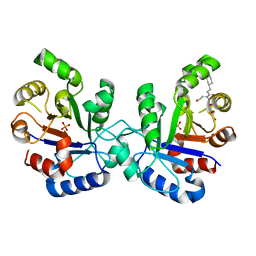 | | Kinetics and structural properties of triosephosphate isomerase from Helicobacter pylori | | Descriptor: | 1-[(3-CYCLOHEXYLPROPANOYL)(2-HYDROXYETHYL)AMINO]-1-DEOXY-D-ALLITOL, PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Chu, C.-H, Lai, Y.-J, Sun, Y.-J. | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetics and Structural Properties of Triosephosphate Isomerase from Helicobacter Pylori
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2KFW
 
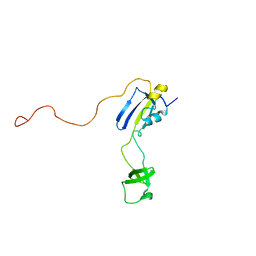 | | Solution structure of full-length SlyD from E.coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase slyD | | Authors: | Martino, L, He, Y, Hands-Taylor, K.L, Valentine, E.R, Kelly, G, Giancola, C, Conte, M.R. | | Deposit date: | 2009-02-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The interaction of the Escherichia coli protein SlyD with nickel ions illuminates the mechanism of regulation of its peptidyl-prolyl isomerase activity.
Febs J., 276, 2009
|
|
7L7J
 
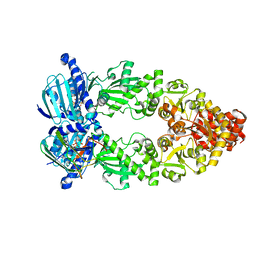 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7L7S
 
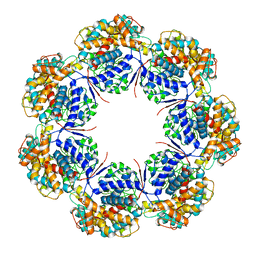 | | Human mitochondrial chaperonin mHsp60 | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Chen, L, Wang, J.C.Y. | | Deposit date: | 2020-12-30 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the structural dynamics of human mitochondrial chaperonin mHsp60.
Sci Rep, 11, 2021
|
|
2WFI
 
 | |
3ZI4
 
 | |
1YYA
 
 | |
1ZOL
 
 | | native beta-PGM | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Zhang, G, Tremblay, L.W, Dai, J, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic cycling in beta-phosphoglucomutase: a kinetic and structural analysis
Biochemistry, 44, 2005
|
|
1ZJA
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 (triclinic form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Trehalulose synthase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2005-04-28 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
1ZMF
 
 | | C domain of human cyclophilin-33(hcyp33) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Wang, T, Yun, C.-H, Gu, S.-Y, Chang, W.-R, Liang, D.-C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88A crystal structure of the C domain of hCyP33: A novel domain of peptidyl-prolyl cis-trans isomerase
Biochem.Biophys.Res.Commun., 333, 2005
|
|
1ZUW
 
 | | Crystal structure of B.subtilis glutamate racemase (RacE) with D-Glu | | Descriptor: | D-GLUTAMIC ACID, glutamate racemase 1 | | Authors: | Ruzheinikov, S.N, Taal, M.A, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-Induced Conformational Changes in Bacillus subtilis Glutamate Racemase and Their Implications for Drug Discovery
Structure, 13, 2005
|
|
1ZZG
 
 | |
4WJE
 
 | |
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8Z38
 
 | | FK506 binding protein 1B (including FK506) | | Descriptor: | 6-CARBOXYPIPERIDINE, peptidylprolyl isomerase | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-resolution cocrystallized crystal structures of FK506 and FK506-binding proteins
To Be Published
|
|
8YO3
 
 | |
8YO9
 
 | |
7OML
 
 | |
4Y9A
 
 | |
4YCC
 
 | |
4YCJ
 
 | | Structure of Acetobacter aceti PurE Y154F | | Descriptor: | 1,2-ETHANEDIOL, N5-carboxyaminoimidazole ribonucleotide mutase, SULFATE ION | | Authors: | Kappock, T.J, Sullivan, K.L. | | Deposit date: | 2015-02-20 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure of a single tryptophan mutant of Acetobacter aceti PurE
To Be Published
|
|
