7JSQ
 
 | | Refined structure of the C-terminal domain of DNAJB6b | | Descriptor: | DnaJ homolog subfamily B member 6 | | Authors: | Karamanos, T.K, Clore, G.M. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An S/T motif controls reversible oligomerization of the Hsp40 chaperone DNAJB6b through subtle reorganization of a beta sheet backbone.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6URP
 
 | |
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
6QES
 
 | | [1-40]Gga-AvBD11 | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M8O
 
 | | Platelet integrin alfaIIb-beta3 cytoplasmic domain | | Descriptor: | platelet integrin alfaIIb subunit: cytoplasmic domain, platelet integrin beta3 subunit: cytoplasmic domain | | Authors: | Vinogradova, O, Velyvis, A, Velyviene, A, Hu, B, Haas, T, Plow, E.F, Qin, J. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Structural mechanism of integrin alfaiib beta3
"Inside-out" Activation as regulated by its cytoplasmic face.
Cell(Cambridge,Mass.), 110, 2002
|
|
1W1F
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-06-17 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Lyn-SH3 Domain in Complex with a Herpesviral Protein Reveals an Extended Recognition Motif that Enhances Binding Affinity.
Protein Sci., 14, 2005
|
|
6Q5Z
 
 | | H-Vc7.2, H-superfamily conotoxin | | Descriptor: | Conotoxin Vc7.2 | | Authors: | Nielsen, L.D, Foged, M.M, Teilum, K, Ellgaard, L. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework.
J.Biol.Chem., 294, 2019
|
|
1W4J
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family.
J. Mol. Biol., 353, 2005
|
|
6QET
 
 | | [41-82]Gga-AvBD11 | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QEU
 
 | | Gga-AvBD11 (Avian beta-defensin 11 from Gallus gallus) | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1MWY
 
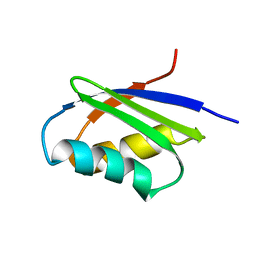 | | Solution structure of the N-terminal domain of ZntA in the apo-form | | Descriptor: | ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1MWZ
 
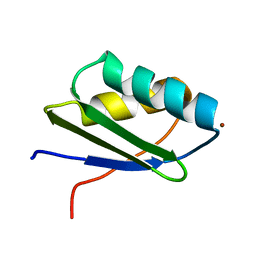 | | Solution structure of the N-terminal domain of ZntA in the Zn(II)-form | | Descriptor: | ZINC ION, ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
8GDH
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7BQE
 
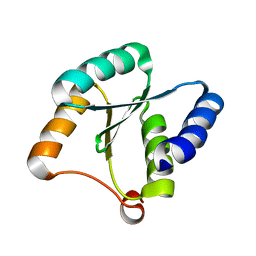 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
1MKN
 
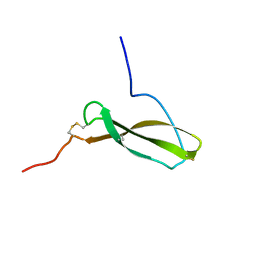 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | Descriptor: | NF6 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
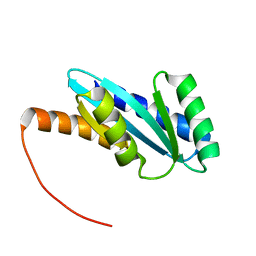 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
1MC7
 
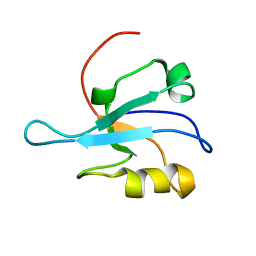 | | Solution Structure of mDvl1 PDZ domain | | Descriptor: | Segment polarity protein dishevelled homolog DVL-1 | | Authors: | Wong, H.-C, Bourdelas, A, Shao, Y, Wu, D, Shi, D.L, Zheng, J. | | Deposit date: | 2002-08-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled
Mol.Cell, 12, 2003
|
|
1MNB
 
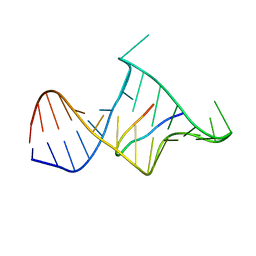 | | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIV TAR RNA, BIV TAT PEPTIDE | | Authors: | Puglisi, J.D, Chen, L, Blanchard, S, Frankel, A.D. | | Deposit date: | 1996-07-25 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex.
Science, 270, 1995
|
|
7MSV
 
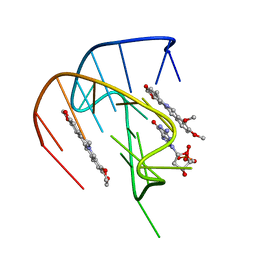 | |
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
3W6I
 
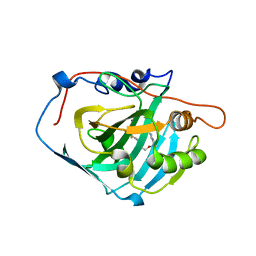 | | Crystal structure of 19F probe-labeled hCAI | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
7AC1
 
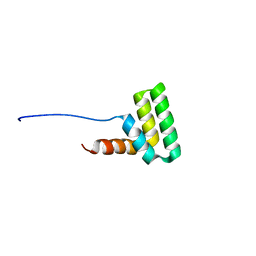 | | Solution structure of the TAF4-RST domain | | Descriptor: | Transcription initiation factor TFIID subunit 4 | | Authors: | Staby, L, Davidsen, R, Bugge, K, Skriver, K, Kragelund, B.B. | | Deposit date: | 2020-09-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | alpha alpha-hub coregulator structure and flexibility determine transcription factor binding and selection in regulatory interactomes.
J.Biol.Chem., 298, 2022
|
|
