5QDF
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000295a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}2-(1~{H}-benzimidazol-2-yl)benzene-1,2-diamine | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4PKS
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5QE0
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000648a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(1,2-oxazole-5-carbonyl)amino]benzoic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEG
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000278a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}1-(4,6-dimethylpyrimidin-2-yl)benzene-1,4-diamine | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5TM3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,3-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | (1S)-2,3-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5QEX
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000123a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4PLH
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, SODIUM ION, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
5TMV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS analog, 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5QFR
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000497a | | Descriptor: | (1S,4R,5S,6R)-2-(methylsulfonyl)-2-azabicyclo[3.3.1]nonane-4,6-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.622 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5TN5
 
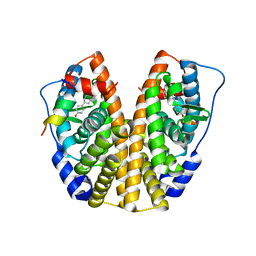 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the AC-ring estrogen, (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4AGS
 
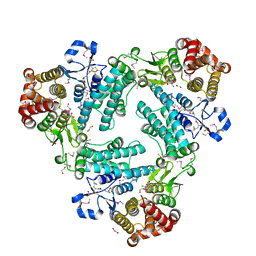 | | Leishmania TDR1 - a unique trimeric glutathione transferase | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, THIOL-DEPENDENT REDUCTASE 1 | | Authors: | Fyfe, P.K, Westrop, G.D, Silva, A.M, Coombs, G.H, Hunter, W.N. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leishmania Tdr1 Structure, a Unique Trimeric Glutathione Transferase Capable of Deglutathionylation and Antimonial Prodrug Activation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DTV
 
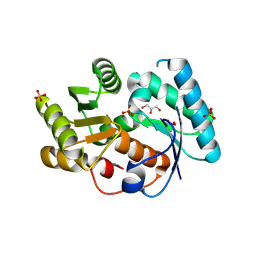 | | Crystal structure of arylmalonate decarboxylase | | Descriptor: | Arylmalonate decarboxylase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Nakasako, M, Obata, R, Miyamaoto, K, Ohta, H. | | Deposit date: | 2008-07-16 | | Release date: | 2009-07-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inverting the Enantioselectivity of Arylmalonate Decarboxylase Revealed by the Structural Analysis of the Gly74Cys/Cys188Ser Mutant in the Liganded Form
Biochemistry, 49, 2010
|
|
2WXP
 
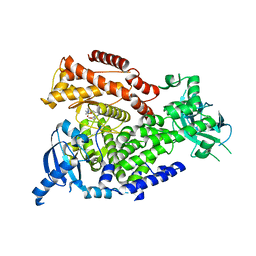 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with GDC-0941. | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
1OMW
 
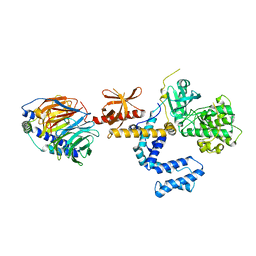 | | Crystal Structure of the complex between G Protein-Coupled Receptor Kinase 2 and Heterotrimeric G Protein beta 1 and gamma 2 subunits | | Descriptor: | G-protein coupled receptor kinase 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1 | | Authors: | Lodowski, D.T, Pitcher, J.A, Capel, W.D, Lefkowitz, R.J, Tesmer, J.J.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Keeping G proteins at Bay: A Complex Between G Protein-Coupled Receptor Kinase 2 and G-Beta-Gamma
Science, 300, 2003
|
|
5T7B
 
 | |
2AL6
 
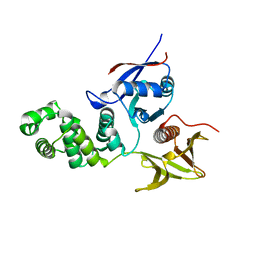 | | FERM domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
3K7C
 
 | |
5TCJ
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate and BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5D5Y
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3K97
 
 | |
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
5YA1
 
 | | crystal structure of LsrK-HPr complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
2WW2
 
 | | Structure of the Family GH92 Inverting Mannosidase BT2199 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1S-8AB-OCTAHYDRO-INDOLIZIDINE-1A,2A,8B-TRIOL, ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
7FMC
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06B07 from the F2X-Universal Library | | Descriptor: | 1-[(2S)-2-hydroxy-3-(methylamino)propyl]pyrrolidin-2-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3RPV
 
 | | CDK2 in complex with inhibitor RC-2-88 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(3-aminobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-27 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
