1ZE7
 
 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in water solution at pH 6.5 | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
3K0Y
 
 | |
6M9U
 
 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
4OCC
 
 | | co-crystal structure of MDM2(17-111) in complex with compound 48 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
3WDS
 
 | |
4ODF
 
 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 47 | | Descriptor: | 6-{[(2S,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2006 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
3WVW
 
 | | Crystal structure of RuCO/apo-WTFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Fujita, K, Tanaka, Y, Abe, S, Hikage, T, Kuchimaru, T, Kizaka-Kondoh, S, Ueno, T. | | Deposit date: | 2014-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular CO release from composite of ferritin and ruthenium carbonyl complexes.
J.Am.Chem.Soc., 136, 2014
|
|
3CBZ
 
 | |
3WFG
 
 | | Mineralocorticoid receptor ligand-binding domain with compuond 2e | | Descriptor: | 1,2-ETHANEDIOL, 6-[(2S)-4-(4-fluorophenyl)-2-methyl-5-oxo-2,5-dihydrofuran-3-yl]-2H-1,4-benzoxazin-3(4H)-one, Mineralocorticoid receptor | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and structure-activity relationships of dihydrofuran-2-one and dihydropyrrol-2-one derivatives as novel benzoxazin-3-one-based mineralocorticoid receptor antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4G26
 
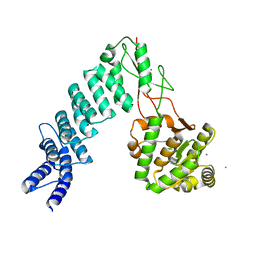 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Ca | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1SCN
 
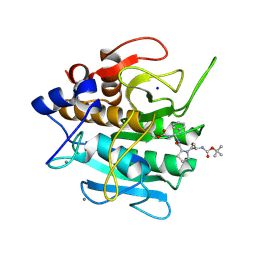 | | INACTIVATION OF SUBTILISIN CARLSBERG BY N-(TERT-BUTOXYCARBONYL-ALANYL-PROLYL-PHENYLALANYL)-O-BENZOL HYDROXYLAMINE: FORMATION OF COVALENT ENZYME-INHIBITOR LINKAGE IN THE FORM OF A CARBAMATE DERIVATIVE | | Descriptor: | CALCIUM ION, N-(tert-butoxycarbonyl)-L-alanyl-N-[(1R)-1-(carboxyamino)-2-phenylethyl]-L-prolinamide, SODIUM ION, ... | | Authors: | Steinmetz, A.C.U, Demuth, H.-U, Ringe, D. | | Deposit date: | 1994-03-02 | | Release date: | 1994-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inactivation of subtilisin Carlsberg by N-((tert-butoxycarbonyl)alanylprolylphenylalanyl)-O-benzolhydroxyl- amine: formation of a covalent enzyme-inhibitor linkage in the form of a carbamate derivative.
Biochemistry, 33, 1994
|
|
4GF9
 
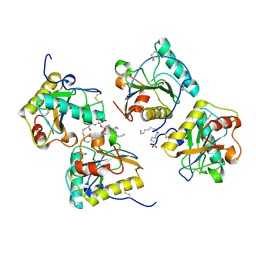 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
1OS0
 
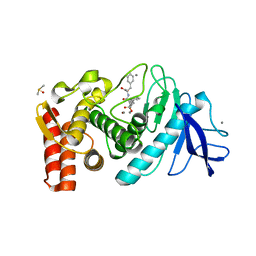 | | Thermolysin with an alpha-amino phosphinic inhibitor | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N-{(2R)-3-[(S)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Selkti, M, Tomas, A, Prange, T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of a new alpha-aminophosphinic derivative inside the active site of TLN (thermolysin): a model for zinc-metalloendopeptidase inhibition.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4C6W
 
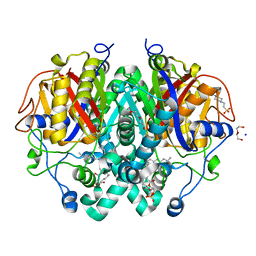 | | Crystal structure of M. tuberculosis C171Q KasA | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(10R)-10-methyloctadecanoyl]oxy}propyl phosphate, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Schiebel, J, Kapilashrami, K, Fekete, A, Bommineni, G.R, Schaefer, C.M, Mueller, M.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Recognition of Mycolic Acid Precursors by Kasa, a Condensing Enzyme and Drug Target from Mycobacterium Tuberculosis
J.Biol.Chem., 288, 2013
|
|
1U7N
 
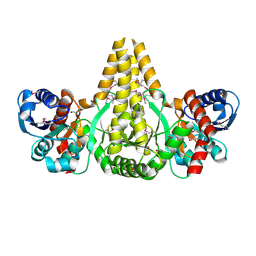 | | Crystal Structure of the fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Fatty acid/phospholipid synthesis protein plsX | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
1YXY
 
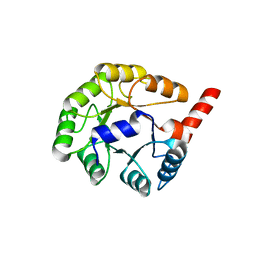 | | Crystal Structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes (APC29713) Structural genomics, MCSG | | Descriptor: | Putative N-acetylmannosamine-6-phosphate 2-epimerase | | Authors: | Rotella, F.J, Zhang, R.G, Lezondra, L.E.O, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes
To be Published
|
|
3O1K
 
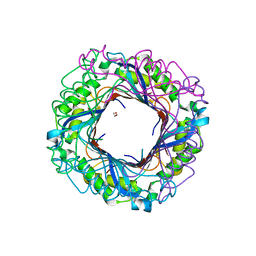 | | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Dihydroneopterin aldolase FolB, putative | | Authors: | Nocek, B, Zhou, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
4HWM
 
 | |
4C6M
 
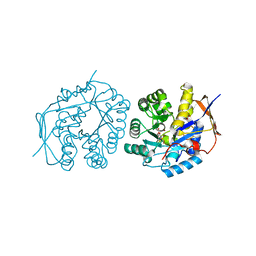 | | Crystal structure of the dihydroorotase domain of human CAD bound to the inhibitor fluoroorotate at pH 7.0 | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
1P4A
 
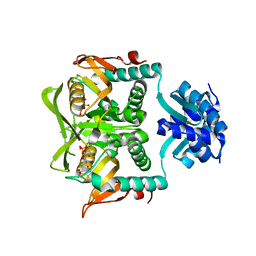 | | Crystal Structure of the PurR complexed with cPRPP | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, Pur operon repressor | | Authors: | Bera, A.K, Zhu, J, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional dissection of the Bacillus subtilis pur operator site.
J.Bacteriol., 185, 2003
|
|
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4GIV
 
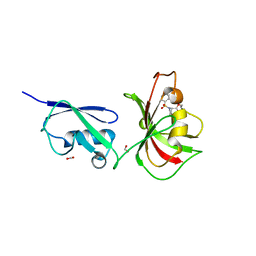 | |
3CET
 
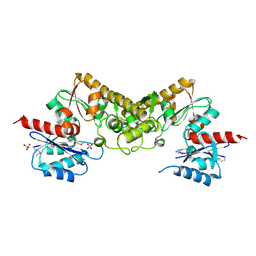 | | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | Conserved archaeal protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, J.F, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
To be Published
|
|
2CYD
 
 | | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, LITHIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae
To be Published
|
|
1T7M
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxYF motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
