8YEL
 
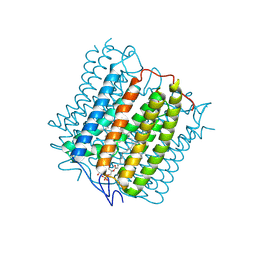 | | Cryo-EM structure of the channelrhodopsin GtCCR4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cation channel rhodopsin 4, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4
Mol.Cell, 2024
|
|
8YEK
 
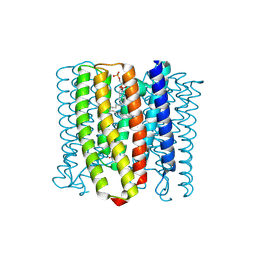 | | Cryo-EM structure of the channelrhodopsin GtCCR2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4
Mol.Cell, 2024
|
|
8YEJ
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 focused on the monomer | | Descriptor: | GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4
Mol.Cell, 2024
|
|
8YE9
 
 | |
8YE6
 
 | |
8YE0
 
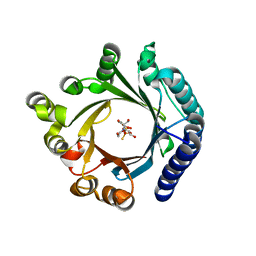 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8YD3
 
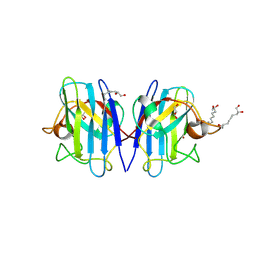 | | Crystal structure of human Cu-Zn Superoxide Dismutase 1 in complex with 1,2,10-Decanetriol | | Descriptor: | (2S)-decane-1,2,10-triol, GLYCEROL, Superoxide dismutase [Cu-Zn], ... | | Authors: | Aouti, S, Padmanabhan, B. | | Deposit date: | 2024-02-19 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of human Cu-Zn Superoxide Dismutase 1 in complex with 1,2,10-Decanetriol
To Be Published
|
|
8YBK
 
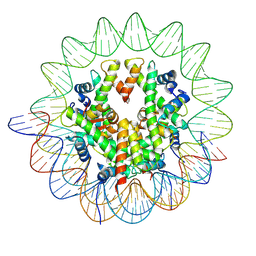 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
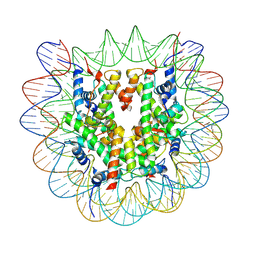 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBF
 
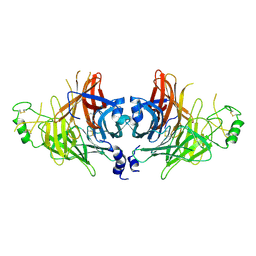 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
8YB7
 
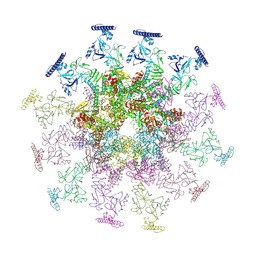 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C3 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 2024
|
|
8YB5
 
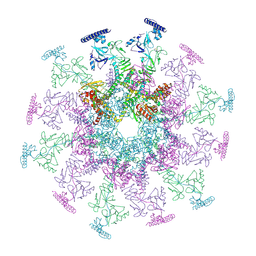 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 2024
|
|
8YAX
 
 | |
8YAN
 
 | |
8YAM
 
 | |
8Y9X
 
 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
8Y9P
 
 | |
8Y95
 
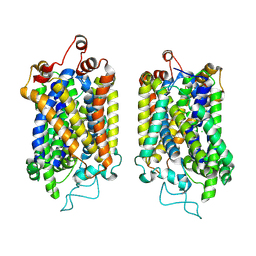 | | Structure of NET-NE in Occluded state | | Descriptor: | CHLORIDE ION, Noradrenaline, SODIUM ION, ... | | Authors: | Zhang, H, Xu, H.E, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y94
 
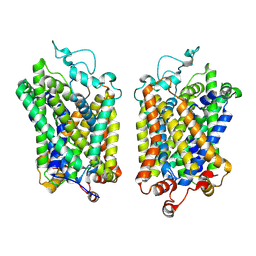 | |
8Y93
 
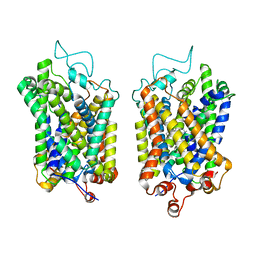 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y92
 
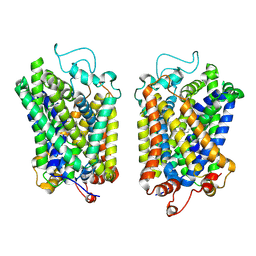 | | structure of NET-Atomoxetine in outward-open state | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y91
 
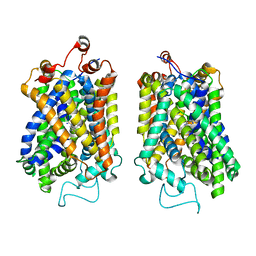 | | Structure of NET-nomifensine in outward-open state | | Descriptor: | (4S)-2-methyl-4-phenyl-3,4-dihydro-1H-isoquinolin-8-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y90
 
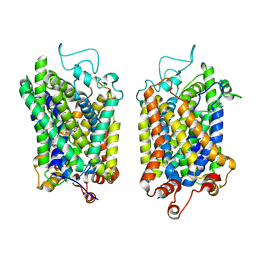 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y8Z
 
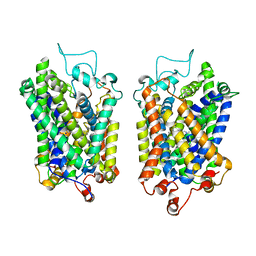 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y8Y
 
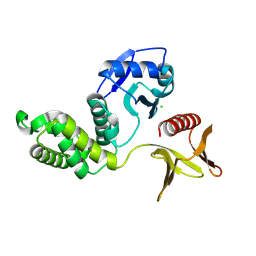 | | Crystal structure of the PTPN21 FERM domain | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Lee, H.S, Ku, B. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of the FERM domain of human protein tyrosine phosphatase non-receptor type 21.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
