3SVE
 
 | |
3SS0
 
 | |
3SSH
 
 | |
3SSK
 
 | |
3SVC
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: chloride complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3SSL
 
 | |
3SSY
 
 | |
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3SSP
 
 | |
3SSV
 
 | |
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
5Y8R
 
 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
3EVP
 
 | | crystal structure of circular-permutated EGFP | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3CFA
 
 | |
3GEX
 
 | |
1BFP
 
 | | BLUE VARIANT OF GREEN FLUORESCENT PROTEIN | | Descriptor: | BLUE FLUORESCENT PROTEIN | | Authors: | Wachter, R.M, Remington, S.J. | | Deposit date: | 1997-04-09 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein.
Biochemistry, 36, 1997
|
|
7A7L
 
 | | rsEGFP in the green-off state | | Descriptor: | Green fluorescent protein, TETRAETHYLENE GLYCOL | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A7K
 
 | | rsEGFP in the green-on state | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZUI
 
 | | Crystal structure of the Cys-Ser mutant of the cpYFP-based biosensor for hypochlorous acid | | Descriptor: | HTH-type transcriptional repressor NemR,Green fluorescent protein,Green fluorescent protein,HTH-type transcriptional repressor NemR | | Authors: | Tossounian, M.A, Van Molle, I, Messens, J. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.200082 Å) | | Cite: | Hypocrates is a genetically encoded fluorescent biosensor for (pseudo)hypohalous acids and their derivatives.
Nat Commun, 13, 2022
|
|
7AA5
 
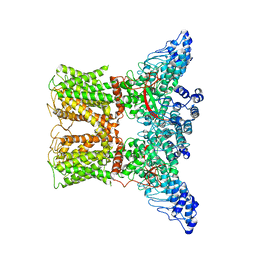 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
6ZSM
 
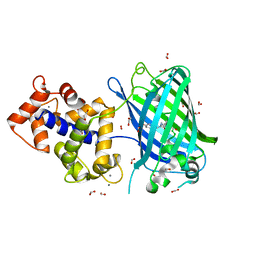 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the ON state (non-illuminated) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
6ZSN
 
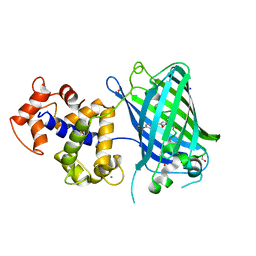 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Green fluorescent protein,Calmodulin, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
7CAO
 
 | | Crystal structure of red chromoprotein from Olindias formosa | | Descriptor: | Chromoprotein | | Authors: | Nakashima, R, Zhai, L, Ike, Y, Matsudz, T, Nagai, T. | | Deposit date: | 2020-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based analysis and evolution of a monomerized red-colored chromoprotein from the Olindias formosa jellyfish.
Protein Sci., 31, 2022
|
|
4Q9W
 
 | |
