2XYJ
 
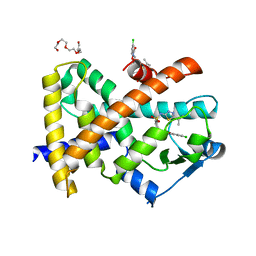 | |
3IZX
 
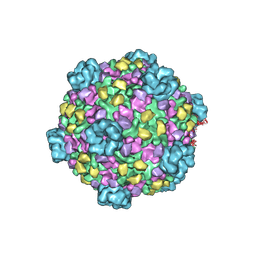 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
1MQI
 
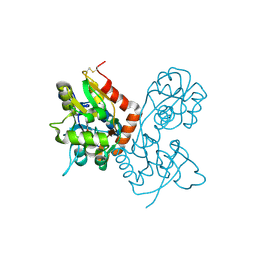 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
4D73
 
 | | X-ray structure of a peroxiredoxin | | Descriptor: | 1-CYS PEROXIREDOXIN | | Authors: | Staudacher, V, Djuika, C.F, Koduka, J, Schlossarek, S, Kopp, J, Buechler, M, Lanzer, M, Deponte, M. | | Deposit date: | 2014-11-19 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasmodium Falciparum Antioxidant Protein Reveals a Novel Mechanism for Balancing Turnover and Inactivation of Peroxiredoxins
Free Radic.Biol.Med., 85, 2015
|
|
9DFX
 
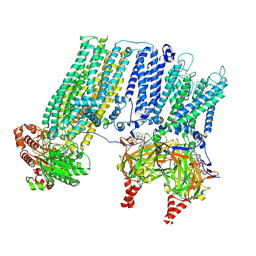 | | Cryo-EM structure of a SUR1/Kir6.2 ATP-sensitive potassium channel in the presence of Aekatperone in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Driggers, C.M, ElSheikh, A, Shyng, S.-L. | | Deposit date: | 2024-08-30 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | AI-based discovery and cryoEM structural elucidation of a K ATP channel pharmacochaperone.
Elife, 13, 2025
|
|
8DUK
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-(methylamino)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-{4-[2-(methylamino)ethoxy]phenyl}-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
5N8C
 
 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5AKE
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5S74
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
2XVL
 
 | | crystal structure of alpha-xylosidase (GH31) from Cellvibrio japonicus in complex with Pentaerythritol propoxylate (5 4 PO OH) | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, ALPHA-XYLOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Izumi, A, Ibatullin, F, Nakhai, A, Gilbert, H.J, Davies, G.J, Brumer, H. | | Deposit date: | 2010-10-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Enzymatic Characterisation of a Glycoside Hydrolase Family 31 Alpha-Xylosidase from Cellvibrio Japonicus Involved in Xyloglucan Saccharification.
Biochem.J., 436, 2011
|
|
6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
5S48
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1741982125 | | Descriptor: | 1~{H}-pyridin-2-one, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1JPT
 
 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
6RI5
 
 | |
5NAT
 
 | |
3EC4
 
 | |
5S8P
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with E07179c (space group P212121) | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | XChem group deposition
To Be Published
|
|
2Y54
 
 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Fragment 1) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
4B74
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-[(2-ammonioethyl)amino]-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function
Nat.Chem.Biol., 8, 2012
|
|
3HEM
 
 | |
6TWF
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
4QSK
 
 | | Crystal Structure of L. monocytogenes Pyruvate Carboxylase in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CITRATE ANION, MANGANESE (II) ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function.
Cell(Cambridge,Mass.), 158, 2014
|
|
4QTP
 
 | | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Anti-sigma factor antagonist, CITRIC ACID, ... | | Authors: | Dranow, D.M, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis
TO BE PUBLISHED
|
|
5S8O
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N01460c (space group P212121) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-(4-chloro-2-methylphenyl)acetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | XChem group deposition
To Be Published
|
|
