1RXP
 
 | |
3CPR
 
 | |
1BBD
 
 | |
2O3M
 
 | | Monomeric G-DNA tetraplex from human C-kit promoter | | Descriptor: | 5'-D(*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*CP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Burge, S, Neidle, S, Patel, D.J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter.
J.Am.Chem.Soc., 129, 2007
|
|
1AX9
 
 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, LAUE DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Ravelli, R.B.G, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-11-03 | | Release date: | 1998-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3D4L
 
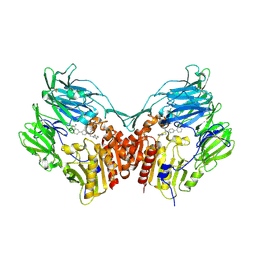 | | Human dipeptidyl peptidase IV/CD26 in complex with a novel inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of new binding elements in DPP-4 inhibition and their applications in novel DPP-4 inhibitor design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3Q4C
 
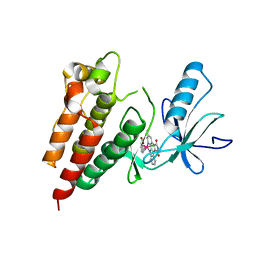 | | Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292 | | Descriptor: | Serine/threonine-protein kinase B-raf, [(1,2,3,4,5,6-eta)-(1S,2R,3R,4R,5S,6S)-1-carboxycyclohexane-1,2,3,4,5,6-hexayl](chloro)(3-methyl-5,7-dioxo-6,7-dihydro-5H-pyrido[2,3-a]pyrrolo[3,4-c]carbazol-12-ide-kappa~2~N~1~,N~12~)ruthenium(1+) | | Authors: | Xie, P, Streu, C, Qin, J, Pregman, H, Pagano, N, Meggers, E, Marmorstein, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase.
Biochemistry, 48, 2009
|
|
3PCI
 
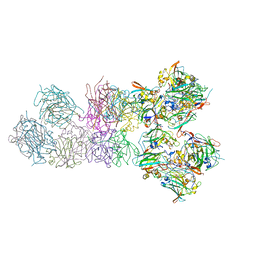 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-IODO-4-HYDROXYBENZOATE | | Descriptor: | 3-IODO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCH
 
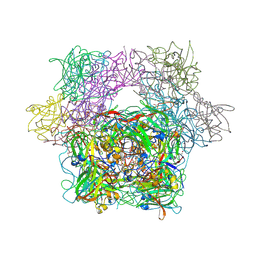 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-CHLORO-4-HYDROXYBENZOATE | | Descriptor: | 3-CHLORO-4-HYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-01-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
1B2U
 
 | |
2YAK
 
 | | Structure of death-associated protein Kinase 1 (dapk1) in complex with a ruthenium octasporine ligand (OSV) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, RUTHENIUM OCTASPORINE 4 | | Authors: | Feng, L, Geisselbrecht, Y, Blanck, S, Wilbuer, A, Atilla-Gokcumen, G.E, Filippakopoulos, P, Kraeling, K, Celik, M.A, Harms, K, Maksimoska, J, Marmorstein, R, Frenking, G, Knapp, S, Essen, L.-O, Meggers, E. | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structurally Sophisticated Octahedral Metal Complexes as Highly Selective Protein Kinase Inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
3D62
 
 | | Development of Broad-Spectrum Halomethyl Ketone Inhibitors Against Coronavirus Main Protease 3CLpro | | Descriptor: | 3C-like proteinase, benzyl (2-oxopropyl)carbamate | | Authors: | Bacha, U, Barrila, J, Gabelli, S.B, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2008-05-18 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CL(pro).
Chem.Biol.Drug Des., 72, 2008
|
|
2YKE
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[(4R)-4-(3H-imidazo[4,5-c]pyridin-2-yl)-4H-fluoren-9-yl]quinoline-5-carboxamide | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
3Q5U
 
 | |
2DRR
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
3D7H
 
 | |
3M8P
 
 | | HIV-1 RT with NNRTI TMC-125 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2010-03-18 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3Q6T
 
 | | Salivary protein from Lutzomyia longipalpis, Ligand free | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
2DSX
 
 | | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68 A resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Chen, C.-J, Lin, Y.-H, Huang, Y.-C, Liu, M.-Y. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.68 Å) | | Cite: | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68A resolution
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1B3C
 
 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | Descriptor: | PROTEIN (NEUROTOXIN CSE-I) | | Authors: | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | Deposit date: | 1998-12-08 | | Release date: | 1998-12-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
2YJX
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-(3H-imidazo[4,5-c]pyridin-2-yl)-3,4-dihydropyrido[2,1-a]isoindol-6(2H)-one, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-24 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2DRS
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263S mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
1NPS
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
2YK2
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-(3H-imidazo[4,5-c]pyridin-2-yl)-3,4-dihydropyrido[2,1-a]isoindol-6(2H)-one, 4-(5-METHYL-4-PHENYLISOXAZOL-3-YL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-25 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
