6LEI
 
 | |
7C3Y
 
 | | Crystal structure of the N-terminal domain of human MdmX protein in complex with Nutlin3a | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Su, Z.D, Cheng, X.Y, Zhang, B.L, Kuang, Z.K, Yang, J, Li, Z.C, Yu, J.P, Zhao, Z.T, Cao, C.Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Crystal structure of the N-terminal domain of human MdmX protein in complex with Nutlin3a
To Be Published
|
|
6LFX
 
 | |
5CTI
 
 | | Crystal structure of the type IX collagen NC2 hetero-trimerization domain with a guest fragment a2a1a1 of type I collagen (native form) | | Descriptor: | Collagen alpha-1(I) chain,Collagen alpha-1(IX) chain, Collagen alpha-1(I) chain,Collagen alpha-3(IX) chain, Collagen alpha-2(I) chain,Collagen alpha-2(IX) chain, ... | | Authors: | Boudko, S.P, Bachinger, H.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8994 Å) | | Cite: | Structural insight for chain selection and stagger control in collagen.
Sci Rep, 6, 2016
|
|
1BJR
 
 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
4NCY
 
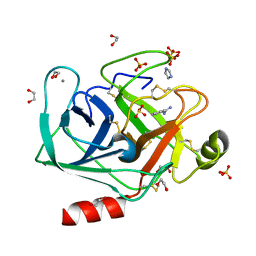 | | In situ trypsin crystallized on a MiTeGen micromesh with imidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WY1
 
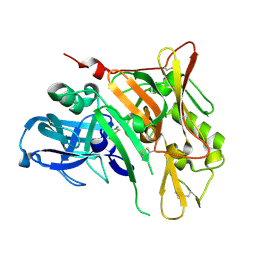 | | Crystal structure of human BACE-1 bound to Compound 24B | | Descriptor: | (4aR,8aS)-8a-(2,4-difluorophenyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1 | | Authors: | Vajdos, F.F, Parris, K. | | Deposit date: | 2014-11-14 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
2A7U
 
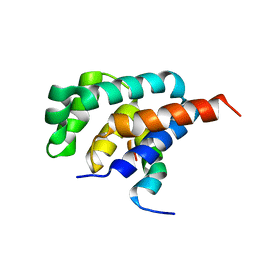 | | NMR solution structure of the E.coli F-ATPase delta subunit N-terminal domain in complex with alpha subunit N-terminal 22 residues | | Descriptor: | ATP synthase alpha chain, ATP synthase delta chain | | Authors: | Wilkens, S, Borchardt, D, Weber, J, Senior, A.E. | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Interaction of the delta and alpha Subunits of the Escherichia coli F(1)F(0)-ATP Synthase by NMR Spectroscopy
Biochemistry, 44, 2005
|
|
7EXK
 
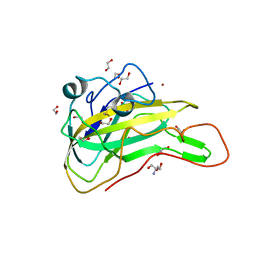 | | An AA9 LPMO of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
6KYJ
 
 | | Hybrid-Rubisco (rice RbcL and sorghum RbcS) in complex with sulfate ions | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, ... | | Authors: | Matsumura, H, Yoshizawa, T, Tanaka, S, Yoshikawa, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C 4 Plant-like High Catalytic Activity.
Mol Plant, 13, 2020
|
|
4X7E
 
 | |
6KZN
 
 | | Crystal Structure Of VIM-2 Metallo-beta-lactamase In Complex With Inhibitor X2 | | Descriptor: | 2,5-diethyl-1-methyl-4-sulfamoyl-pyrrole-3-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
1BRS
 
 | |
8JUW
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
1BTX
 
 | |
4X00
 
 | |
6F4W
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
7COV
 
 | | Potato D-enzyme, native (substrate free) | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, CALCIUM ION, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6UOL
 
 | |
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
4NG3
 
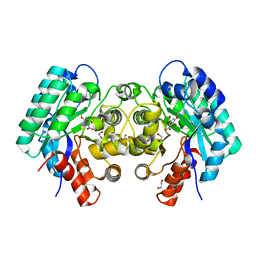 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid
To be Published
|
|
5OA3
 
 | | Human 40S-eIF2D-re-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Weisser, M, Schaefer, T, Leibundgut, M, Boehringer, D, Aylett, C.H.S, Ban, N. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and Functional Insights into Human Re-initiation Complexes.
Mol. Cell, 67, 2017
|
|
6L8L
 
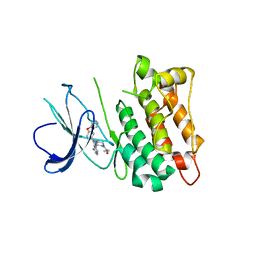 | | C-Src in complex with ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Characterization of ibrutinib as a non-covalent inhibitor of SRC-family kinases.
Bioorg.Med.Chem.Lett., 34, 2020
|
|
5VPX
 
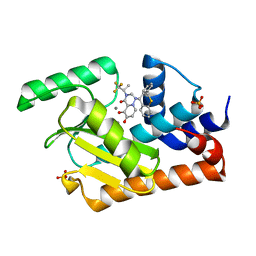 | | I38T mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
