2INB
 
 | |
9B8Q
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, peripheral stalks | | Descriptor: | V-type proton ATPase 116 kDa subunit a isoform 1, V-type proton ATPase subunit C 1, V-type proton ATPase subunit E 1, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
2INP
 
 | | Structure of the Phenol Hydroxylase-Regulatory Protein Complex | | Descriptor: | FE (III) ION, Phenol hydroxylase component phL, Phenol hydroxylase component phM, ... | | Authors: | Sazinsky, M.S, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
5K9C
 
 | | Crystal structure of human dihydroorotate dehydrogenase with ML390 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
9BBI
 
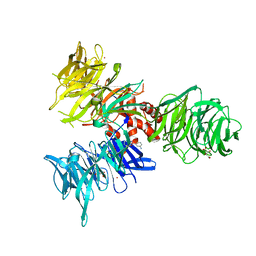 | | Co-crystal structure of human DDB1 bound to fragment UB028669 | | Descriptor: | 3-([1,3]oxazolo[4,5-b]pyridin-2-yl)aniline, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028669
To be published
|
|
9B3P
 
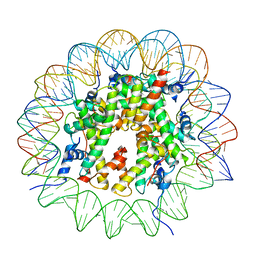 | |
5JPO
 
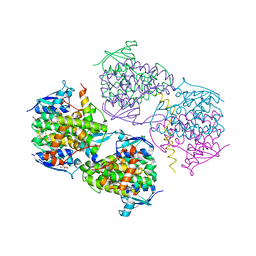 | |
9BFY
 
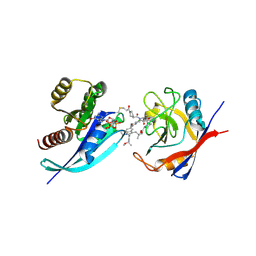 | | Tri-complex of Compound-14, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of Elironrasib (RMC-6291), a Potent and Orally Bioavailable, RAS(ON) G12C-Selective, Covalent Tricomplex Inhibitor for the Treatment of Patients with RAS G12C-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
9BRU
 
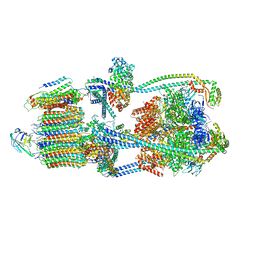 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
5JQV
 
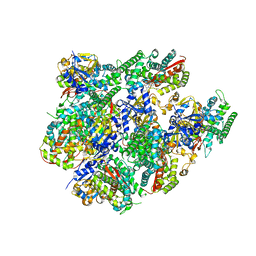 | | Crystal structure of Cytochrome P450 BM3 heme domain T269V/L272W/L322I/A406S (WIVS) variant with iron(III) deuteroporphyrin IX bound | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, FE(III) DEUTEROPORPHYRIN IX | | Authors: | Reynolds, E.W, McHenry, M.W, Cannac, F, Gober, J.G, Snow, C.D, Brustad, E.M. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | An Evolved Orthogonal Enzyme/Cofactor Pair.
J.Am.Chem.Soc., 138, 2016
|
|
2IQA
 
 | | PFA2 FAB fragment, monoclinic apo form | | Descriptor: | ACETAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
9BRT
 
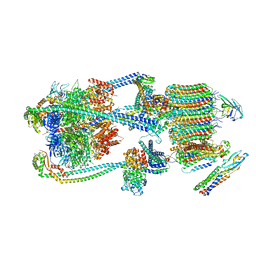 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRB
 
 | |
1LBY
 
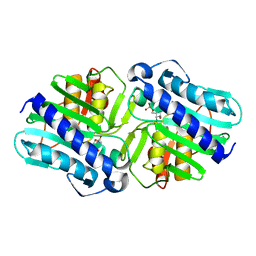 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Manganese ions, Fructose-6-Phosphate, and Phosphate ion | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
9BBG
 
 | | Co-crystal structure of human DDB1 bound to fragment UB028671 | | Descriptor: | 1,2-ETHANEDIOL, 1H-indol-6-amine, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028671
To be published
|
|
2ISY
 
 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
5JRY
 
 | |
9BGP
 
 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, aminotransferase | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
5JQU
 
 | | Crystal structure of Cytochrome P450 BM3 heme domain G265F/T269V/L272W/L322I/F405M/A406S (WIVS-FM) variant with iron(III) deuteroporphyrin IX bound | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, FE(III) DEUTEROPORPHYRIN IX | | Authors: | Reynolds, E.W, McHenry, M.W, Cannac, F, Gober, J.G, Snow, C.D, Brustad, E.M. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | An Evolved Orthogonal Enzyme/Cofactor Pair.
J.Am.Chem.Soc., 138, 2016
|
|
9BRS
 
 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BAF
 
 | | Solution NMR structure of conofurin-Delta | | Descriptor: | Alpha-conotoxin LvIA | | Authors: | Harvey, P.J, Craik, D.J, Hone, A.J, McIntosh, J.M. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Novel Peptide Derivatives of the Severe Acute Respiratory Syndrome-Coronavirus-2 Spike-Protein that Potently Inhibit Nicotinic Acetylcholine Receptors.
J.Med.Chem., 67, 2024
|
|
5JTD
 
 | |
9BBB
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, cobicistat | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with the inhibitor cobicistat: Structural and mechanistic insights and comparison with ritonavir.
Arch.Biochem.Biophys., 758, 2024
|
|
9BC3
 
 | | Transglutaminase 2 - Alternate state | | Descriptor: | CALCIUM ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Mathews, I.I, Sewa, A.S, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
