9VSR
 
 | | The DCY1040 and TUG-891-bound structure of TMEM175 | | Descriptor: | (2S)-2-butyl-2-cyclohexyl-6,7-bis(fluoranyl)-5-[3-(1H-1,2,3,4-tetrazol-5-yl)propoxy]-3H-inden-1-one, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, CHOLESTEROL, ... | | Authors: | Zhu, X, Liu, H, Yin, W. | | Deposit date: | 2025-07-09 | | Release date: | 2025-09-17 | | Last modified: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into the activation of TMEM175 by small molecule.
Neuron, 113, 2025
|
|
9N77
 
 | | SSU processome maturation and disassembly, State K | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9N6V
 
 | | SSU processome maturation and disassembly, State A | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9VNX
 
 | | Crystal Structure of the mutant ROR gamma LBD With Compound 28 | | Descriptor: | (2~{R})-~{N}-[5-(5-cyano-6-propan-2-yloxy-pyridin-3-yl)-1-(trifluoromethyl)pyrazol-3-yl]-1-ethanoyl-4-propan-2-ylsulfonyl-piperazine-2-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Akai, S, Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2025-07-01 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Concise and Modular Approach to Generate Novel ROR gamma Agonists.
J.Med.Chem., 68, 2025
|
|
9N76
 
 | | SSU processome maturation and disassembly, State J | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9B65
 
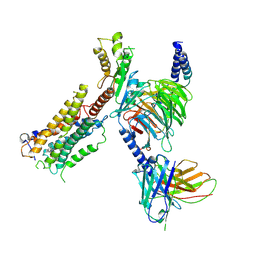 | | Biased agonist bound CB1-Gi structure | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rangari, V.A, O'Brien, E.S, Kobilka, B.K, Krishna Kumar, K, Majumdar, S. | | Deposit date: | 2024-03-23 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A cryptic pocket in CB1 drives peripheral and functional selectivity.
Nature, 640, 2025
|
|
9N6Y
 
 | | SSU processome maturation and disassembly, State C | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
4XJ0
 
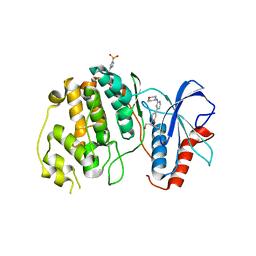 | | Crystal structure of ERK2 in complex with an inhibitor 14K | | Descriptor: | 1-[(1S)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2015-01-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of highly potent, selective, and efficacious small molecule inhibitors of ERK1/2.
J.Med.Chem., 58, 2015
|
|
9N75
 
 | | SSU processome maturation and disassembly, State I | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9RKE
 
 | | Crystal Structure of compound 1-mediated ternary complex of KRAS G12R GCP with pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CITRATE ANION, Elongin-B, ... | | Authors: | Wijaya, A.J, Karolak, N.K, Ciulli, A. | | Deposit date: | 2025-06-13 | | Release date: | 2025-11-12 | | Last modified: | 2025-11-26 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Identification of a Highly Cooperative PROTAC Degrader Targeting GTP-Loaded KRAS(On) Alleles.
J.Am.Chem.Soc., 147, 2025
|
|
5EK0
 
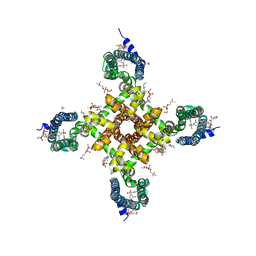 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
2YFC
 
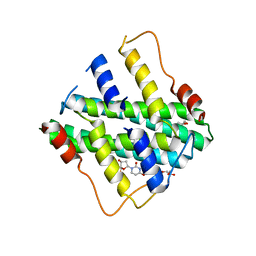 | | STRUCTURAL AND FUNCTIONAL INSIGHTS OF DR2231 PROTEIN, THE MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE FROM DEINOCOCCUS RADIODURANS, COMPLEXED WITH Mn and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Goncalves, A.M.D, De Sanctis, D, Mcsweeney, S.M. | | Deposit date: | 2011-04-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
7AAK
 
 | | Human porphobilinogen deaminase R173W mutant crystallized in the ES2 intermediate state | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
4MC3
 
 | | Hedycaryol synthase in complex with Nerolidol | | Descriptor: | (3R,6E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
9N6X
 
 | | SSU processome maturation and disassembly, State B | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9YE4
 
 | | Crystal structure of the human DCAF1 WDR domain in complex with OICR-41110 | | Descriptor: | (4P)-4-(4-chloro-2-fluorophenyl)-N-[(1S)-1-(3-chloro-4-fluorophenyl)-2-cyanoethyl]-5-[(1E)-3-oxo-3-(piperidin-1-yl)prop-1-en-1-yl]-1H-pyrrole-3-carboxamide, 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1 | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Mamai, A, Al-awar, R, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-09-23 | | Release date: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the human DCAF1 WDR domain in complex with OICR-41110
To be published
|
|
4EHI
 
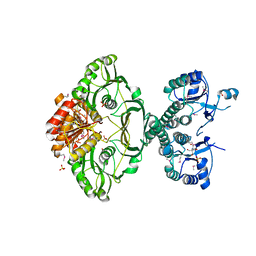 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
6CKS
 
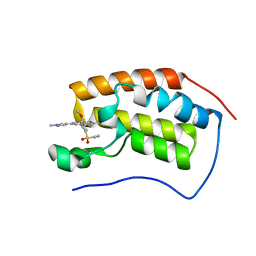 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | 4-[5-(ethylsulfonyl)-2-methoxyphenyl]-2-methyl-6-(1-methyl-1H-pyrazol-4-yl)isoquinolin-1(2H)-one, Bromodomain-containing protein 4 | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2UUV
 
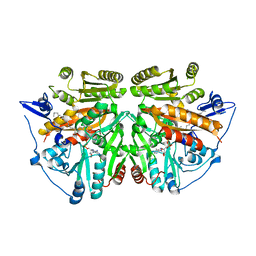 | | alkyldihydroxyacetonephosphate synthase in P1 | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, FLAVIN-ADENINE DINUCLEOTIDE, HEXADECAN-1-OL | | Authors: | Razeto, A, Mattiroli, F, Carpanelli, E, Aliverti, A, Pandini, V, Coda, A, Mattevi, A. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Structure, 15, 2007
|
|
7KRY
 
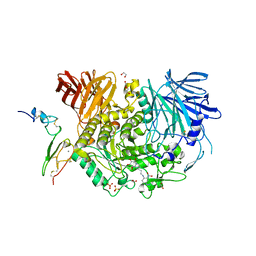 | | Co-crystal structure of alpha glucosidase with compound 11 | | Descriptor: | (1S,2S,3R,4S,5S)-5-({6-[(4-azido-2-nitrophenyl)amino]hexyl}amino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
9O9V
 
 | |
7TBE
 
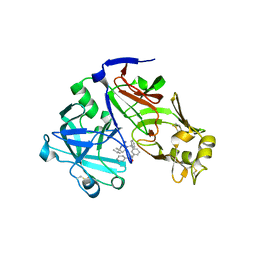 | | Crystal structure of Plasmepsin X from Plasmodium vivax in complex with WM4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(R)-[(2E,4S)-2-imino-4-methyl-6-oxo-4-(propan-2-yl)-1,3-diazinan-1-yl](phenyl)methyl]-N-[(1S)-1-phenylethyl]benzamide, Plasmepsin X, ... | | Authors: | Hodder, A.N, Christensen, J.B, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
4PV7
 
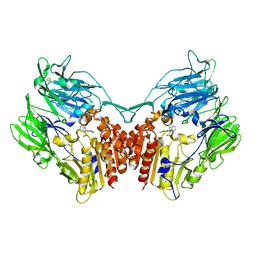 | | Cocrystal structure of dipeptidyl-peptidase 4 with an indole scaffold inhibitor | | Descriptor: | 1-[2-(2,4-dichlorophenyl)-1-(methylsulfonyl)-1H-indol-3-yl]methanamine, Dipeptidyl peptidase 4 soluble form | | Authors: | Xiao, P, Guo, R, Huang, S, Cui, H, Ye, S, Zhang, Z. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery of dipeptidyl peptidase IV (DPP4) inhibitors based on a novel indole scaffold
Chin.Chem.Lett., 25, 2014
|
|
7KUZ
 
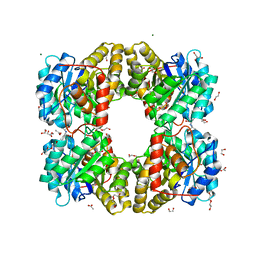 | |
3H3T
 
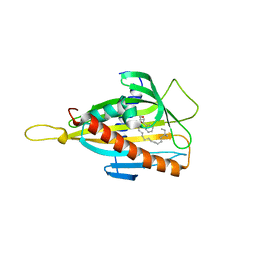 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
