5HO8
 
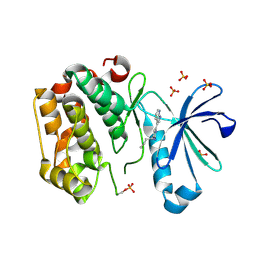 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-butyl-6-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8TXZ
 
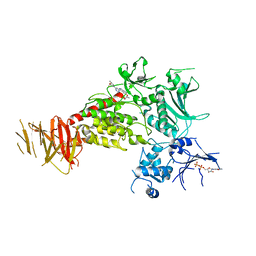 | | Structure of C-terminal LRRK2 bound to MLi-2 | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Sanz-Murillo, M, Villagran-Suarez, A, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
6V0U
 
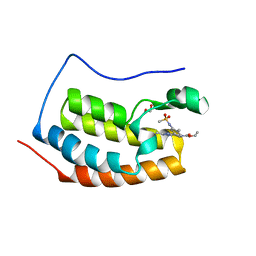 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
7W10
 
 | | UGT74AN2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Long, F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7AZV
 
 | |
7W1B
 
 | | Glycosyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
4O9G
 
 | | Crystal structure of the H51N mutant of the 3,4-ketoisomerase QdtA from Thermoanaerobacterium thermosaccharolyticum in complex with TDP-4-keto-6-deoxyglucose | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, 1,2-ETHANEDIOL, QdtA, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
6EIQ
 
 | | DYRK1A in complex with XMD14-124 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, [4-azanyl-2-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-1,3-thiazol-5-yl]-phenyl-methanone | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7B0E
 
 | | Crystal structure of SmbA loop deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structure of loop deletion mutant SmbA bound to a monomeric c-di-GMP
To Be Published
|
|
5HWJ
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase | | Descriptor: | FORMIC ACID, GLYCEROL, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
7AYW
 
 | |
5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
6MH8
 
 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6V8N
 
 | |
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
4OIN
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6CAX
 
 | |
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
1BPL
 
 | | GLYCOSYLTRANSFERASE | | Descriptor: | ALPHA-1,4-GLUCAN-4-GLUCANOHYDROLASE | | Authors: | Machius, M, Wiegand, G, Huber, R. | | Deposit date: | 1995-07-13 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of calcium-depleted Bacillus licheniformis alpha-amylase at 2.2 A resolution.
J.Mol.Biol., 246, 1995
|
|
3MFD
 
 | | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, D-alanyl-D-alanine carboxypeptidase dacB | | Authors: | Cuff, M.E, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis.
TO BE PUBLISHED
|
|
8TF2
 
 | |
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
9J90
 
 | | Crystal structure of Pseudomonas aeruginosa SuhB complexed with Gallic acid in orthorhombic space group | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 3,4,5-trihydroxybenzoic acid, Nus factor SuhB | | Authors: | Yadav, V.K, Shukla, M, Maji, S, Bhattacharyya, S. | | Deposit date: | 2024-08-21 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa SuhB complexed with Gallic acid in orthorhombic space group
To Be Published
|
|
5K8G
 
 | |
6VFD
 
 | | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site.
To be Published
|
|
