1MSN
 
 | | The HIV protease (mutant Q7K L33I L63I V82F I84V) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MRX
 
 | | Structure of HIV protease (Mutant Q7K L33I L63I V82F I84V ) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
4OF4
 
 | | X-ray structure of unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-14 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.4 A resolution
To be Published
|
|
6CDR
 
 | | Human CtBP1 (28-378) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
2VPO
 
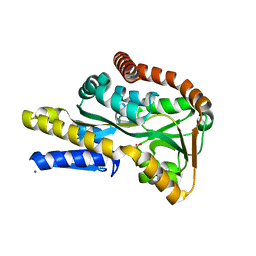 | | High resolution structure of the periplasmic binding protein TeaA from TeaABC TRAP transporter of Halomonas elongata in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
4CDJ
 
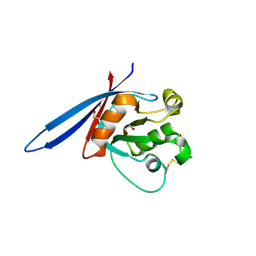 | | Structure of ZNRF3 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE ZNRF3, FORMIC ACID | | Authors: | Peng, W.C, de Lau, W, Madoori, P.K, Forneris, F, Granneman, J.C.M, Clevers, H, Gros, P. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Wnt-Antagonist Znrf3 and its Complex with R-Spondin 1 and Implications for Signaling.
Plos One, 8, 2013
|
|
4O84
 
 | |
5BWV
 
 | |
1MRW
 
 | | Structure of HIV protease (Mutant Q7K L33I L63I) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
4FMZ
 
 | |
4OCJ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SODIUM ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCU
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SULFATE ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1MSM
 
 | | The HIV protease (mutant Q7K L33I L63I) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
4BZV
 
 | | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4BZU
 
 | | The Solution Structure of the MLN 944-d(TATGCATA)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4OCQ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
3NUF
 
 | |
5Z5D
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 | | Descriptor: | Beta-xylosidase, CALCIUM ION, GLYCEROL | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
8GPI
 
 | | HIV-1 Env X18 UFO in complex with 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, ... | | Authors: | Niu, J, Xu, Y.W, Yang, B. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs.
Nat Commun, 14, 2023
|
|
3KC0
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 10b | | Descriptor: | Fructose-1,6-bisphosphatase 1, [(8H-indeno[1,2-d][1,3]thiazol-4-yloxy)methyl]phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2R43
 
 | | I50V HIV-1 protease in complex with an amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
3KJ0
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
2QE9
 
 | |
4YWQ
 
 | | Crystal structure of the ROQ domain of human Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ROQ domain of human Roquin-1
To be Published
|
|
