7W37
 
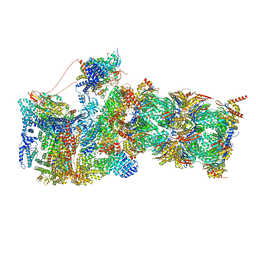 | | Structure of USP14-bound human 26S proteasome in state EA1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3H
 
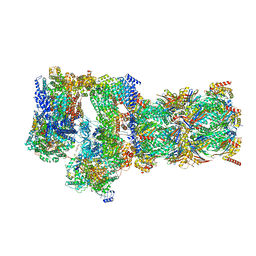 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3G
 
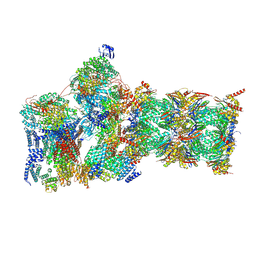 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W39
 
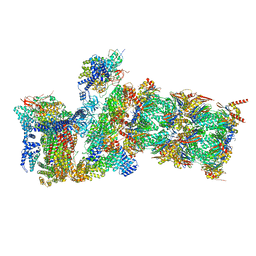 | | Structure of USP14-bound human 26S proteasome in state EA2.1_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3C
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W38
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.0_UBL | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3A
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3J
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3K
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7MHS
 
 | | Structure of p97 (subunits A to E) with substrate engaged | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, Y, Han, H, Cooney, I, Hill, C.P, Shen, P.S. | | Deposit date: | 2021-04-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Active conformation of the p97-p47 unfoldase complex.
Nat Commun, 13, 2022
|
|
7W3M
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3I
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SB_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7QO4
 
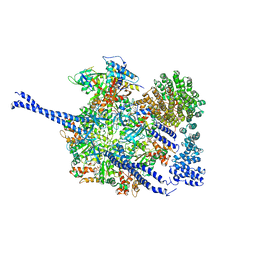 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7WI4
 
 | | Cryo-EM structure of E.Coli FtsH protease cytosolic domains | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7WI3
 
 | | Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Modulator of FtsH protease HflK | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7UPR
 
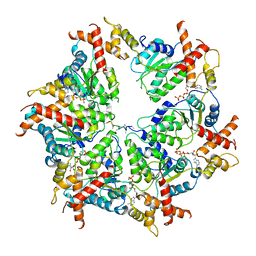 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (closed conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Outer mitochondrial transmembrane helix translocase, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7UPT
 
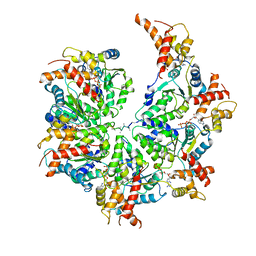 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7SUK
 
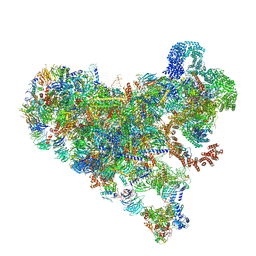 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7U1A
 
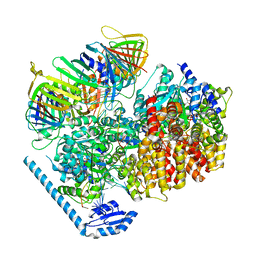 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U1P
 
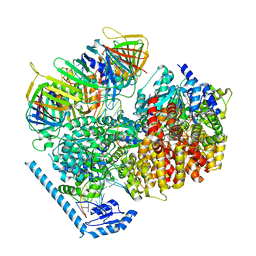 | | RFC:PCNA bound to DNA with a ssDNA gap of five nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U19
 
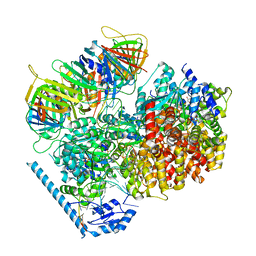 | | RFC:PCNA bound to nicked DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA, MAGNESIUM ION, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7OAT
 
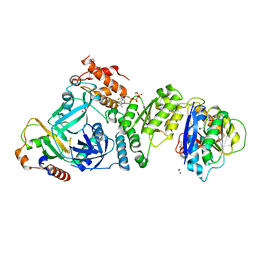 | | Structural basis for targeted p97 remodelling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D6X
 
 | |
8D6Y
 
 | |
8DR3
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
