6DHR
 
 | |
6D8Y
 
 | |
6D8R
 
 | |
6D8U
 
 | |
6D93
 
 | |
6D9P
 
 | |
6D3T
 
 | |
6D8S
 
 | |
6BL9
 
 | |
6CJD
 
 | |
1JSP
 
 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
6D9O
 
 | |
1Q5L
 
 | | NMR structure of the substrate binding domain of DnaK bound to the peptide NRLLLTG | | Descriptor: | Chaperone protein dnaK, peptide NRLLLTG | | Authors: | Stevens, S.Y, Cai, S, Pellecchia, M, Zuiderweg, E.R. | | Deposit date: | 2003-08-08 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG.
Protein Sci., 12, 2003
|
|
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
2ABO
 
 | | NMR structure of gamma herpesvirus 68 a viral Bcl-2 homolog | | Descriptor: | bcl-2 homolog | | Authors: | Loh, J, Huang, Q, Petros, A.M, Nettesheim, D, van Dyk, L.F, Labrada, L, Speck, S.H, Levine, B, Olejniczak, E.T, Virgin, H.W. | | Deposit date: | 2005-07-15 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A surface groove essential for viral Bcl-2 function during chronic infection in vivo.
Plos Pathog., 1, 2005
|
|
1S4W
 
 | | NMR structure of the cytoplasmic domain of integrin AIIb in DPC micelles | | Descriptor: | Integrin alpha-IIb | | Authors: | Vinogradova, O, Vaynberg, J, Kong, X, Haas, T.A, Plow, E.F, Qin, J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane-mediated structural transitions at the cytoplasmic face during integrin activation.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S4X
 
 | | NMR Structure of the integrin B3 cytoplasmic domain in DPC micelles | | Descriptor: | Integrin beta-3 | | Authors: | Vinogradova, O, Vaynberg, J, Kong, X, Haas, T.A, Plow, E.F, Qin, J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane-mediated structural transitions at the cytoplasmic face during integrin activation.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TTK
 
 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
1P94
 
 | | NMR Structure of ParG symmetric dimer | | Descriptor: | plasmid partition protein ParG | | Authors: | Golovanov, A.P, Barilla, D, Golovanova, M, Hayes, F, Lian, L.Y. | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure.
Mol.Microbiol., 50, 2003
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1TT3
 
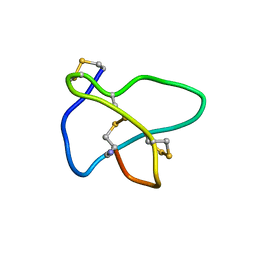 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1MV3
 
 | |
1MUZ
 
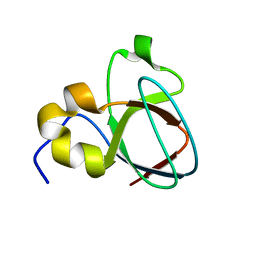 | |
1K3Q
 
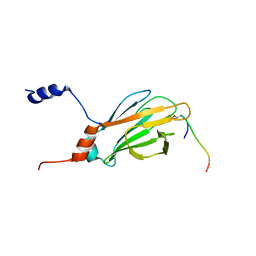 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
