4O6B
 
 | |
2X4R
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to Cytomegalovirus (CMV) pp65 epitope | | Descriptor: | 65 KDA PHOSPHOPROTEIN, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
7YGG
 
 | |
8JRD
 
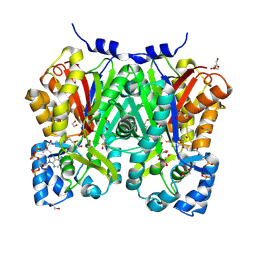 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
7SV6
 
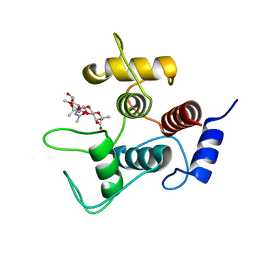 | |
7SV3
 
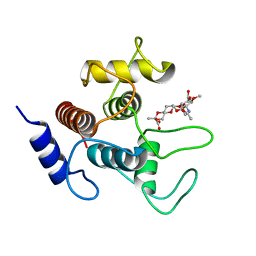 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
6TZV
 
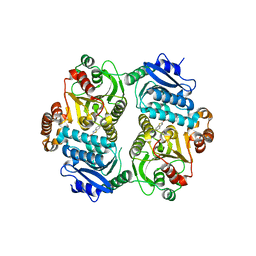 | | Crystal Structure of the carboxyltransferase subunit of ACC (AccD6) in complex with inhibitor Phenyl-Cyclodiaone from Mycobacterium tuberculosis | | Descriptor: | 4'-[(6-chloro-1,3-benzothiazol-2-yl)oxy]-6-hydroxy-4,4-dimethyl-4,5-dihydro[1,1'-biphenyl]-2(3H)-one, Probable propionyl-CoA carboxylase beta chain 6 | | Authors: | Reddy, M.C.M, Zhou, N, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-08-13 | | Release date: | 2020-08-19 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Novel herbicidal derivatives that inhibit carboxyltransferase subunit of the acetyl-CoA carboxylase in Mycobacterium tuberculosis
To Be Published
|
|
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
6FG3
 
 | |
6FKY
 
 | |
8YBC
 
 | |
9H8C
 
 | | Human CDK8/Cyclin-C complex with inhibitor 2-9 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{S})-3-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]piperidin-1-yl]carbonyl-1~{H}-pyrrole-2-carbonitrile, Cyclin-C, ... | | Authors: | Somers, D.O. | | Deposit date: | 2024-10-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Phenotype-Led Identification of IL-10 Upregulators in Human CD4 + T-cells and Elucidation of Their Pharmacology as Highly Selective CDK8/CDK19 Inhibitors.
J.Med.Chem., 68, 2025
|
|
6BXA
 
 | | Crystal structure of N-terminal fragment of Zebrafish Toll-Like Receptor 5 (TLR5) with Lamprey Variable Lymphocyte Receptor 2 (VLR2) bound | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
6GFQ
 
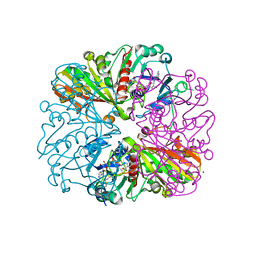 | | cyanobacterial GAPDH with NAD and CP12 bound | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9H8S
 
 | | Human CDK8/Cyclin-C complex with inhibitor 3-7 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{S},5~{R})-3-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]-5-methoxy-piperidin-1-yl]carbonyl-1~{H}-pyrrole-2-carbonitrile, CHLORIDE ION, ... | | Authors: | Somers, D.O. | | Deposit date: | 2024-10-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Phenotype-Led Identification of IL-10 Upregulators in Human CD4 + T-cells and Elucidation of Their Pharmacology as Highly Selective CDK8/CDK19 Inhibitors.
J.Med.Chem., 68, 2025
|
|
7Y5Y
 
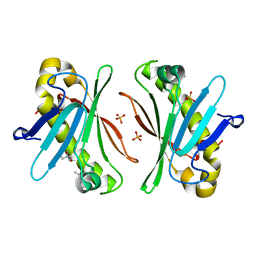 | | X-ray Structure of Stay-Green (SGR) from Anaerolineae bacterium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Dey, D, Nishijima, M, Kurisu, G, Tanaka, H, Ito, H. | | Deposit date: | 2022-06-18 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and reaction mechanism of a bacterial Mg-dechelatase homolog from the Chloroflexi Anaerolineae.
Protein Sci., 31, 2022
|
|
6GI4
 
 | |
8KFU
 
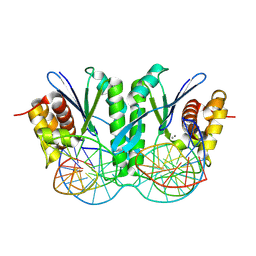 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
5EUQ
 
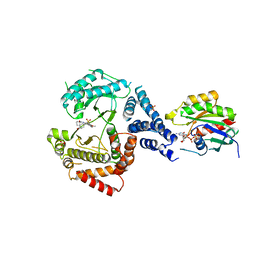 | |
8KFW
 
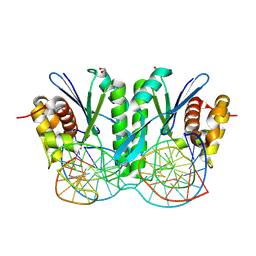 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
6TVB
 
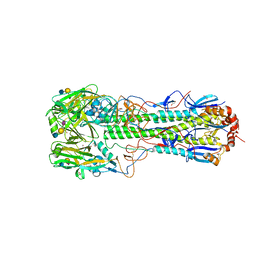 | | Crystal structure of the haemagglutinin from a transmissible H10N7 seal influenza virus isolated in Netherland in complex with human receptor analogue 6'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6C18
 
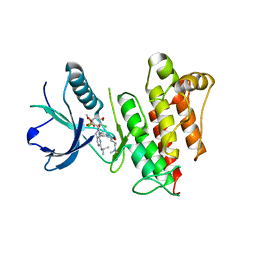 | | FGFR1 kinase complex with inhibitor SN37115 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-[(propan-2-yl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
7YWJ
 
 | | Crystal structure of an engineered TycA variant, TycA pPLA (L313P) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Mittl, P, Camus, A, Truong, G, Markert, G, Hilvert, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Reprogramming Nonribosomal Peptide Synthetases for Site-Specific Insertion of alpha-Hydroxy Acids.
J.Am.Chem.Soc., 144, 2022
|
|
6M8V
 
 | | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex
To Be Published
|
|
