8PP4
 
 | |
8PP5
 
 | |
2SPZ
 
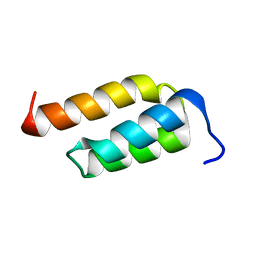 | | STAPHYLOCOCCAL PROTEIN A, Z-DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Montelione, G.T, Tashiro, M, Tejero, R, Lyons, B.A. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the Z domain of staphylococcal protein A.
J.Mol.Biol., 272, 1997
|
|
5AB6
 
 | |
8PP3
 
 | |
5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BSB
 
 | | Lipomyces starkeyi levoglucosan kinase bound to levoglucosan | | Descriptor: | Levoglucosan, Levoglucosan kinase, SULFATE ION | | Authors: | Bacik, J.P. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Producing Glucose 6-Phosphate from Cellulosic Biomass: STRUCTURAL INSIGHTS INTO LEVOGLUCOSAN BIOCONVERSION.
J.Biol.Chem., 290, 2015
|
|
2W70
 
 | |
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
2V20
 
 | | Structure of a TEM-1 beta-lactamase insertant allosterically regulated by kanamycin and anions. Complex with sulfate. | | Descriptor: | BETA-LACTAMASE TEM, SULFATE ION, ZINC ION | | Authors: | Evrard, C, Barrios, H, Mathonet, P, Soumillion, P, Fastrez, J, Declercq, J.P. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Engineering an Allosteric Binding Site for Aminoglycosides Into Tem1-Beta-Lactamase.
Chembiochem, 12, 2011
|
|
7F3X
 
 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
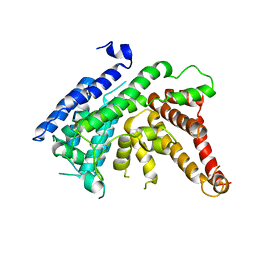 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
2VUX
 
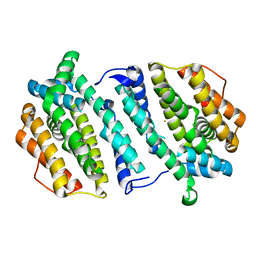 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
2W6Z
 
 | |
2W6O
 
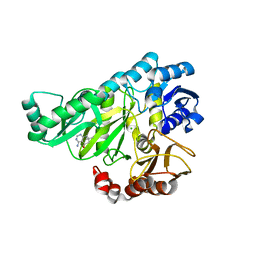 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W9J
 
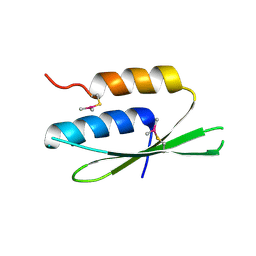 | | The crystal structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle | | Descriptor: | SIGNAL RECOGNITION PARTICLE SUBUNIT SRP14 | | Authors: | Brooks, M.A, Ravelli, R.B.G, McCarthy, A.A, Strub, K, Cusack, S. | | Deposit date: | 2009-01-25 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Srp14 from the Schizosaccharomyces Pombe Signal Recognition Particle.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2X8U
 
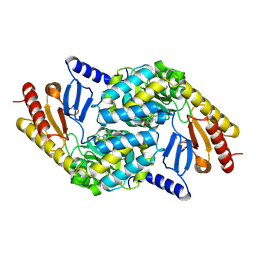 | | Sphingomonas wittichii Serine palmitoyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-03-23 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Serine Palmitoyltransferase from Sphingomonas Wittichii Rw1 an Interesting Link to an Unusual Acyl Carrier Protein
Biopolymers, 93, 2010
|
|
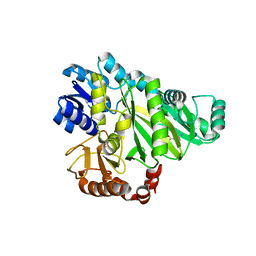
2W6M
 
 | |
2VP8
 
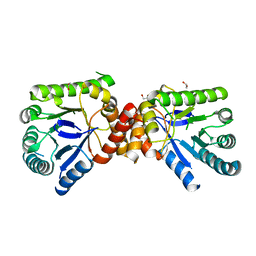 | | Structure of Mycobacterium tuberculosis Rv1207 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPTEROATE SYNTHASE 2 | | Authors: | Gengenbacher, M, Xu, T, Niyomwattanakit, P, Spraggon, G, Dick, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biochemical and Structural Characterization of the Putative Dihydropteroate Synthase Ortholog Rv1207 of Mycobacterium Tuberculosis.
Fems Microbiol.Lett., 287, 2008
|
|
2W6N
 
 | |
7EOE
 
 | | Crystal structure of CCDC25 homodimer | | Descriptor: | Coiled-coil domain-containing protein 25 | | Authors: | Liu, Y.R. | | Deposit date: | 2021-04-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of CCDC25 homodimer
To Be Published
|
|
7EOF
 
 | | Structure of CCDC25-DNA complex | | Descriptor: | Coiled-coil domain-containing protein 25, DNA (5'-D(*CP*AP*GP*AP*TP*CP*AP*CP*TP*AP*GP*TP*AP*GP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*CP*TP*AP*CP*TP*AP*GP*TP*GP*AP*TP*CP*T)-3') | | Authors: | Liu, Y.R. | | Deposit date: | 2021-04-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.733 Å) | | Cite: | Structure of CCDC25-DNA complex
To Be Published
|
|
7F69
 
 | | Crystal structure of WIPI2b in complex with ATG16L1 | | Descriptor: | Isoform 2 of Autophagy-related protein 16-1, Isoform 2 of WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Gong, X.Y, Pan, L.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy.
Sci Adv, 9, 2023
|
|
