7ZE8
 
 | | PucE-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3O6Z
 
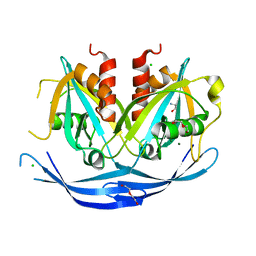 | | Structure of the D152A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
6TBE
 
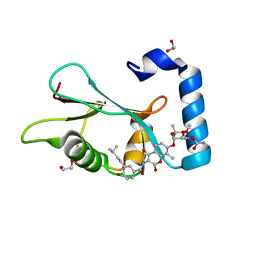 | | LC3A in complex with (3R,4S,5R,6R)-5-hydroxy-6-((4-hydroxy-3-(4-hydroxy-3-isopentylbenzamido)-8-methyl-2-oxo-2H-chromen-7-yl)oxy)-3-methoxy-2,2-dimethyltetrahydro-2H-pyran-4-yl carbamate | | Descriptor: | 1,2-ETHANEDIOL, Microtubule-associated proteins 1A/1B light chain 3A, NOVOBIOCIN | | Authors: | Kramer, J.S, Pogoryelov, D, Hartmann, M, Chaikuad, A, Proschak, E. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67008042 Å) | | Cite: | Demonstrating Ligandability of the LC3A and LC3B Adapter Interface.
J.Med.Chem., 64, 2021
|
|
7PQI
 
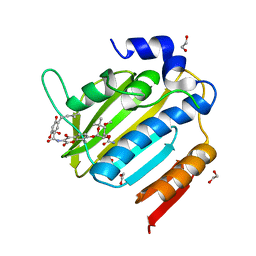 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7CI0
 
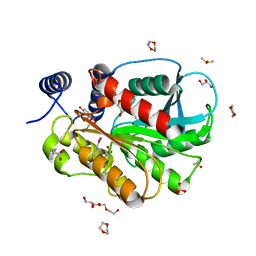 | | Microbial Hormone-sensitive lipase E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7ZCU
 
 | | PucA-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6TI8
 
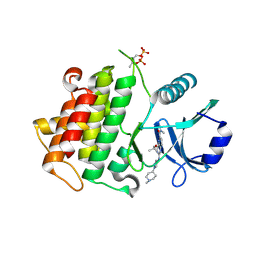 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(1-methylcyclopropyl)oxy-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[3,2-d]pyrimidine-6-carboxamide | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
8RE0
 
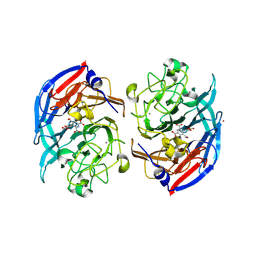 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
7CJX
 
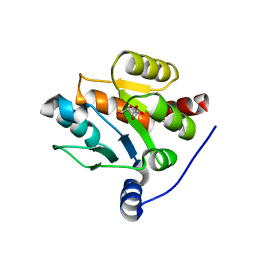 | | UDP-glucuronosyltransferase 2B15 C-terminal domain-L446S | | Descriptor: | L(+)-TARTARIC ACID, UDP-glucuronosyltransferase 2B15 | | Authors: | Wang, C.Y, Zhang, L. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.986414 Å) | | Cite: | Structure of UDP-glucuronosyltransferase 2B15 C-terminal domain L446S at 1.99 Angstroms resolution
To Be Published
|
|
8R3Q
 
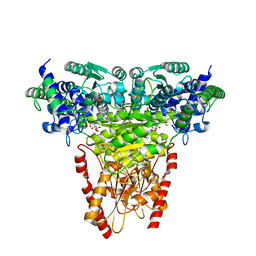 | | Transketolase from Plasmodium falciparum in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Biochemical, Bioinformatic, and Structural Comparisons of Transketolases and Position of Human Transketolase in the Enzyme Evolution.
Biochemistry, 63, 2024
|
|
8HYI
 
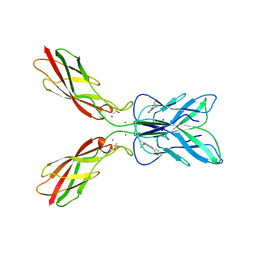 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine | | Descriptor: | 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Modulation of a conformational ensemble by a small molecule that inhibits key protein-protein interactions involved in cell adhesion.
Protein Sci., 32, 2023
|
|
3OBL
 
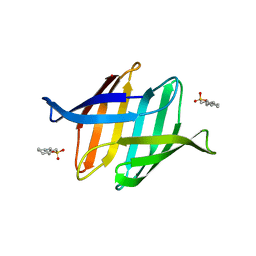 | |
6GKV
 
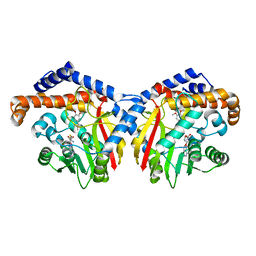 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8J5M
 
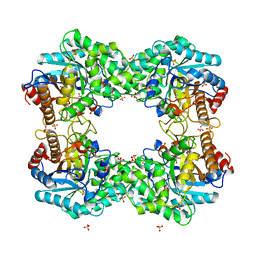 | |
4YBB
 
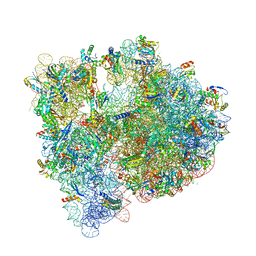 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6TMB
 
 | |
5IXX
 
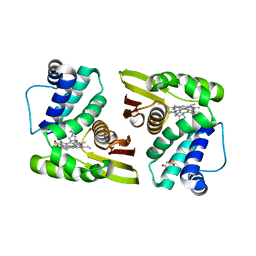 | |
4N1Z
 
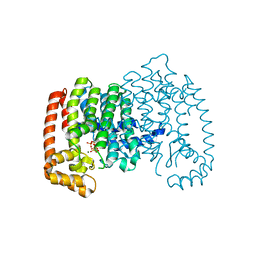 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1222 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Liu, Y.L, Xia, Y, Zhang, Y, Verma, I, Oldfield, E. | | Deposit date: | 2013-10-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin.
Sci Transl Med, 6, 2014
|
|
6SE8
 
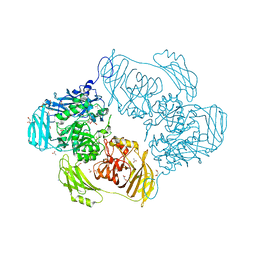 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
7ZSZ
 
 | |
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
6SA2
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX3 (10) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-morpholin-4-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6I5Z
 
 | | Papaver somniferum O-methyltransferase | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE | | Authors: | Davies, G.J, Cabry, M.P, Offen, W.A. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6SB1
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
