2LC4
 
 | | Solution Structure of PilP from Pseudomonas aeruginosa | | Descriptor: | PilP protein | | Authors: | Howell, P, Tammam, S, Chong, P, Forman-Kay, J.D. | | Deposit date: | 2011-04-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the PilN, PilO and PilP type IVa pilus subcomplex.
Mol.Microbiol., 82, 2011
|
|
5IT5
 
 | | Thermus thermophilus PilB core ATPase region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP binding motif-containing protein PilF, MAGNESIUM ION, ... | | Authors: | Mancl, J, Robinson, H, Black, W, Yang, Z, Schubot, F. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Crystal Structure of a Type IV Pilus Assembly ATPase: Insights into the Molecular Mechanism of PilB from Thermus thermophilus.
Structure, 24, 2016
|
|
3FIP
 
 | | Crystal structure of Usher PapC translocation pore | | Descriptor: | Outer membrane usher protein papC | | Authors: | Huang, Y, Deisenhofer, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.154 Å) | | Cite: | Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ITF
 
 | |
3F6L
 
 | | Structure of the F4 fimbrial chaperone FaeE | | Descriptor: | Chaperone protein faeE | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F6I
 
 | | Structure of the SeMet labeled F4 fibrial chaperone FaeE | | Descriptor: | Chaperone protein faeE | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F65
 
 | | The F4 fimbrial chaperone FaeE does not self-cap its interactive surfaces | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chaperone protein faeE, PHOSPHATE ION | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4Z7X
 
 | | MdbA protein, a thiol-disulfide oxidoreductase from Actinomyces oris. | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, MdbA | | Authors: | OSIPIUK, J, Reardon-Robinson, M.E, Ton-That, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris.
J.Biol.Chem., 290, 2015
|
|
7CG9
 
 | |
6BBT
 
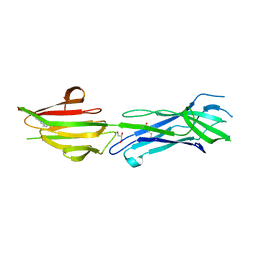 | | Structure of the major pilin protein (T-13) from Streptococcus pyogenes serotype GAS131465 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Young, P.G, Baker, E.N, Moreland, N.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
6BBW
 
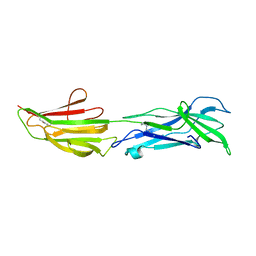 | |
8GMX
 
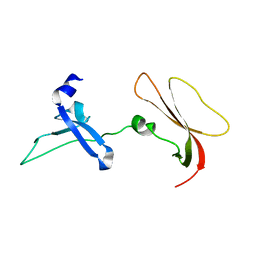 | |
2Y4X
 
 | |
4IPV
 
 | |
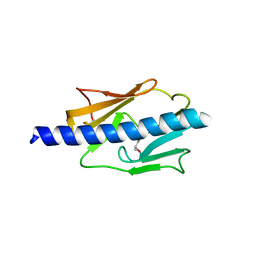
4IPU
 
 | |
4FOK
 
 | | 1.8 A Crystal structure of the FimX EAL domain in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX, GLYCEROL | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-20 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
4FOJ
 
 | | 1.55 A Crystal Structure of Xanthomonas citri FimX EAL domain in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-20 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
5D55
 
 | | Crystal structure of the E. coli Hda pilus minor tip subunit, HdaB | | Descriptor: | CITRATE ANION, HdaB,HdaA (Adhesin), HUS-associated diffuse adherence, ... | | Authors: | Lee, W.-C, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and analysis of HdaB: The enteroaggregative Escherichia coli AAF/IV pilus tip protein.
Protein Sci., 25, 2016
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
2KEP
 
 | | Solution structure of XcpT, the main component of the type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | General secretion pathway protein G | | Authors: | Alphonse, S, Durand, E, Douzi, B, Darbon, H, Filloux, A, Voulhoux, R, Bernard, C. | | Deposit date: | 2009-02-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system
J.Struct.Biol., 2009
|
|
8TWC
 
 | | Acinetobacter phage AP205 T=3 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TW2
 
 | | Acinetobacter phage AP205 T=4 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
5TFY
 
 | |
2EYU
 
 | | The Crystal Structure of the C-terminal Domain of Aquifex aeolicus PilT | | Descriptor: | SULFATE ION, twitching motility protein PilT | | Authors: | Satyshur, K.A, Worzalla, G.A, Meyer, L.S, Heiniger, E.K, Aukema, K.G, Forest, K.T. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility.
Structure, 15, 2007
|
|
8C27
 
 | |
