5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XT6
 
 | | A sulfur-transferring catalytic intermediate of SufS-SufU complex from Bacillus subtilis | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Cysteine desulfurase SufS, ZINC ION, ... | | Authors: | Fujishiro, T, Kunichika, K, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
5XT5
 
 | | SufS-SufU complex from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YNA
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM alpha-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNC
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM beta-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YBW
 
 | | Crystal structure of pyridoxal 5'-phosphate-dependent aspartate racemase | | Descriptor: | Aspartate racemase | | Authors: | Mizobuchi, T, Nonaka, R, Yoshimura, M, Abe, K, Takahashi, S, Kera, Y, Goto, M. | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a pyridoxal 5'-phosphate-dependent aspartate racemase derived from the bivalve mollusc Scapharca broughtonii
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5YKY
 
 | | Crystal Structure of Cross-Linked Tetragonal Hen Egg White Lysozyme Soaked with 10 mM Rose Bengal and 10mM H2PtCl6 | | Descriptor: | CHLORIDE ION, Lysozyme C, PLATINUM (II) ION, ... | | Authors: | Tabe, H, Takahashi, H, Shimoi, T, Abe, S, Ueno, T, Yamada, Y. | | Deposit date: | 2017-10-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Photocatalytic hydrogen evolution systems constructed in cross-linked porous protein crystals
Appl Catal B, 237, 2018
|
|
5YNE
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM alpha-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YPX
 
 | | Crystal structure of calaxin with magnesium | | Descriptor: | Calaxin, MAGNESIUM ION | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-11-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
5YN2
 
 | | Crystal structure of apo Pullulanase from Klebsiella pneumoniae in space group P43212 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PulA protein | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YN7
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 0.1 mM beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YND
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DUF3372 domain-containing protein, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNH
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5ZC9
 
 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
2D17
 
 | | Solution RNA structure of stem-bulge-stem region of the HIV-1 dimerization initiation site | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*AP*GP*AP*GP*GP*CP*GP*AP*CP*CP*C)-3', 5'-R(*GP*GP*GP*UP*CP*GP*GP*CP*UP*UP*GP*CP*UP*G)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1E
 
 | | Crystal structure of PcyA-biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CYX
 
 | | Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) | | Descriptor: | Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Yoshikawa, S, Arai, R, Murayama, K, Imai, Y, Takahashi, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7)
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2D1A
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2D18
 
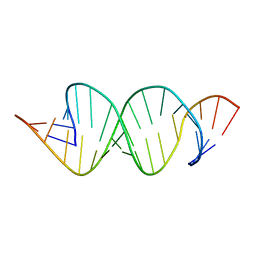 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1B
 
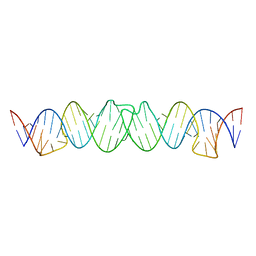 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2DKE
 
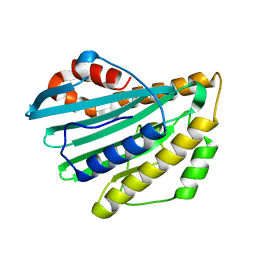 | | Crystal structure of substrate-free form of PcyA | | Descriptor: | CHLORIDE ION, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Sugishima, M, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Induced-fitting and electrostatic potential change of PcyA upon substrate binding demonstrated by the crystal structure of the substrate-free form
Febs Lett., 580, 2006
|
|
