3FGV
 
 | |
3FJ2
 
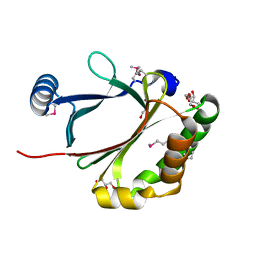 | |
3GZ7
 
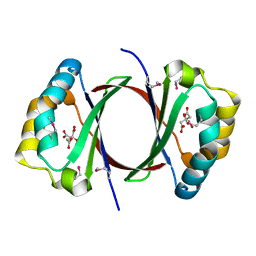 | |
3HX9
 
 | | Structure of heme-degrader, MhuD (Rv3592), from Mycobacterium tuberculosis with two hemes bound in its active site | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Protein Rv3592 | | Authors: | Chim, N, Nguyen, T.Q, Iniguez, A, Goulding, C.W. | | Deposit date: | 2009-06-19 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unusual Diheme Conformation of the Heme-Degrading Protein from Mycobacterium tuberculosis
J.Mol.Biol., 395, 2009
|
|
3KG1
 
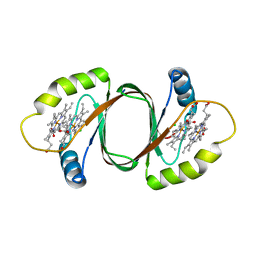
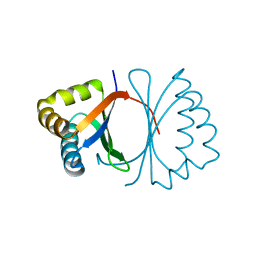 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KKF
 
 | |
3KNG
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3LGM
 
 | |
3LGN
 
 | | Crystal structure of IsdI in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, MAGNESIUM ION, OXYGEN MOLECULE, ... | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2010-01-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The IsdG-family of haem oxygenases degrades haem to a novel chromophore
Mol.Microbiol., 75, 2010
|
|
3MCS
 
 | |
3QGP
 
 | |
3TVZ
 
 | | Structure of Bacillus subtilis HmoB | | Descriptor: | Putative uncharacterized protein yhgC | | Authors: | Choe, J, Choi, S, Park, S. | | Deposit date: | 2011-09-21 | | Release date: | 2012-07-11 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus subtilis HmoB is a heme oxygenase with a novel structure.
Bmb Rep, 45, 2012
|
|
4DN9
 
 | | CRYSTAL STRUCTURE OF putative Antibiotic biosynthesis monooxygenase from Chloroflexus aurantiacus J-10-fl | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Antibiotic biosynthesis monooxygenase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF putative Antibiotic biosynthesis monooxygenase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
4DPO
 
 | | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1 | | Descriptor: | Conserved protein | | Authors: | Agarwal, R, Chamala, S, Evans, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Foti, R, Siedel, R, Zencheck, W, Villigas, G, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-13 | | Release date: | 2012-02-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1
To be Published
|
|
4FNI
 
 | |
4FNH
 
 | | Crystal structure of IsdI-W66Y in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inactivation of the heme degrading enzyme IsdI by an active site substitution that diminishes heme ruffling.
J.Biol.Chem., 287, 2012
|
|
4FVC
 
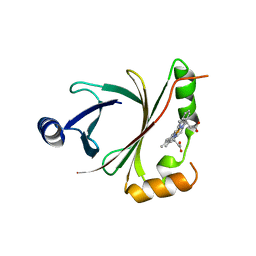 | | HmoB structure with heme | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein yhgC | | Authors: | Park, S. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HmoB with Heme
To be Published
|
|
4HL9
 
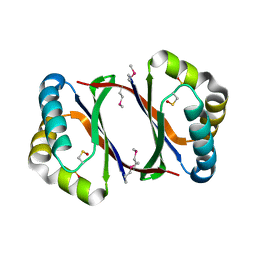 | | Crystal structure of antibiotic biosynthesis monooxygenase | | Descriptor: | Antibiotic biosynthesis monooxygenase | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of antibiotic biosynthesis monooxygenase
To be Published
|
|
4JOU
 
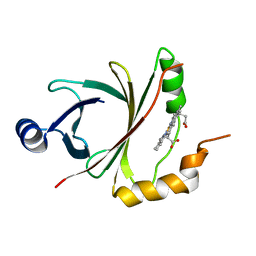 | |
4NL5
 
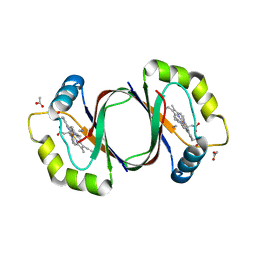 | | Mycobacterium tuberculosis heme-degrading protein MhuD in complex with heme and cyanide | | Descriptor: | ACETATE ION, CYANIDE ION, Heme-degrading monooxygenase HmoB, ... | | Authors: | Morse, R.P, Chao, A, Goulding, C.W. | | Deposit date: | 2013-11-13 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Spectroscopic Insights into Heme Degradation by Mycobacterium tuberculosis MhuD.
Inorg.Chem., 53, 2014
|
|
4NPO
 
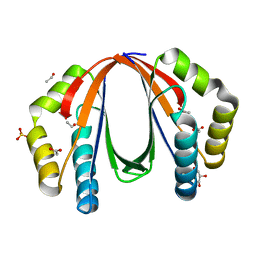 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
4OZ5
 
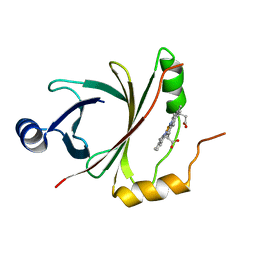 | | Bacillus subtilis HmoB | | Descriptor: | Bacillus subtilis HmoB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Choe, J, Park, S. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Bacillus subtilis HmoB
To Be Published
|
|
4ZOS
 
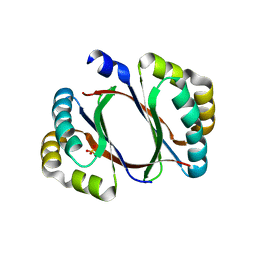 | | 2.20 Angstrom resolution crystal structure of protein YE0340 of unidentified function from Yersinia enterocolitica subsp. enterocolitica 8081] | | Descriptor: | PHOSPHATE ION, protein YE0340 from Yersinia enterocolitica subsp. enterocolitica 8081 | | Authors: | Halavaty, A.S, Wawrzak, A, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20 Angstrom resolution crystal structure of protein YE0340 of unidentified function from Yersinia enterocolitica subsp. enterocolitica 8081]
To Be Published
|
|
5F9P
 
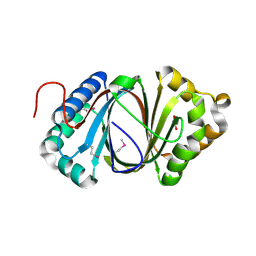 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
