6TRH
 
 | |
6HTY
 
 | | PXR in complex with P2X4 inhibitor compound 25 | | Descriptor: | (2~{R})-~{N}-[4-(3-chloranylphenoxy)-3-sulfamoyl-phenyl]-2-phenyl-propanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V, Werner, S, Mesch, S, Laux-Biehlmann, A, Braeuer, N, Dahloef, H, Klint, J, ter Laak, A, Pook, E, Neagoe, I, Nubbemeyer, R, Schulz, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Characterization of the Potent and Selective P2X4 InhibitorN-[4-(3-Chlorophenoxy)-3-sulfamoylphenyl]-2-phenylacetamide (BAY-1797) and Structure-Guided Amelioration of Its CYP3A4 Induction Profile.
J.Med.Chem., 62, 2019
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YN3
 
 | | Crystal structure of Cucumisin complex with pro-peptide | | Descriptor: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
5JTT
 
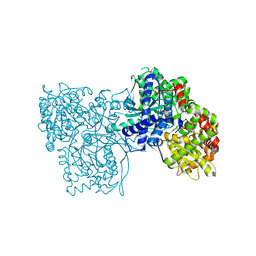 | | Crystal structure of GPb in complex with 8a | | Descriptor: | (1S)-1,5-anhydro-1-(5-phenyl-1H-imidazol-2-yl)-D-glucitol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur.J.Med.Chem., 123, 2016
|
|
6ZRB
 
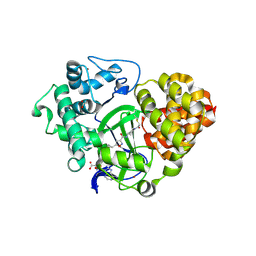 | |
6SHD
 
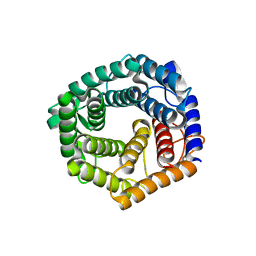 | |
5JV0
 
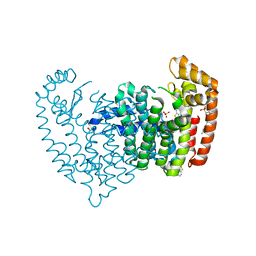 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-038 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-2-(3-fluorophenyl)-1-{[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
3MQD
 
 | |
8TL9
 
 | | Human Type 3 IP3 Receptor - Resting State (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
7TFQ
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
5FGE
 
 | |
8Z2L
 
 | |
8TXN
 
 | |
5JX5
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo- and endo- activity in a complex with cellobiose
Acta Crystallogr.,Sect.D, 2019
|
|
6LX2
 
 | | Potato D-enzyme complexed with CA26 | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, 4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
8Z2T
 
 | | Crystal structure of trehalose synthase from Deinococcus radiodurans complexed with validoxylamine A (VAA) | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Ye, L.C, Chen, S.C. | | Deposit date: | 2024-04-13 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural and Mutational Analyses of Trehalose Synthase from Deinococcus radiodurans Reveal the Interconversion of Maltose-Trehalose Mechanism.
J.Agric.Food Chem., 72, 2024
|
|
7X05
 
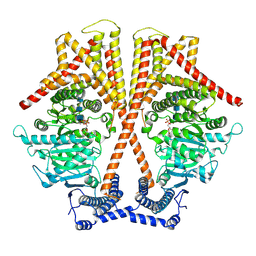 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with the nascent chitooligosaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase, MANGANESE (II) ION, ... | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
8FT7
 
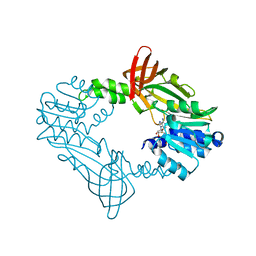 | |
9IVZ
 
 | | Apg mutant enzyme D448A of acarbose hydrolase from human gut flora K. grimontii TD1, complex with acarbose | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-5-[[(1~{S},4~{R},5~{S},6~{S})-3-(hydroxymethyl)-4,5,6-tris(oxidanyl)cyclohex-2-en-1-yl]amino]-6-methyl-oxane-2,3,4-triol-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, Maltodextrin glucosidase | | Authors: | Zhou, J.H, Huang, J.Y. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-30 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular insights of acarbose metabolization catalyzed by acarbose-preferred glucosidase.
Nat Commun, 16, 2025
|
|
8Z7D
 
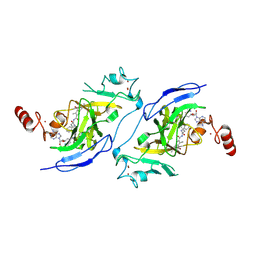 | | Structure of G9a in complex with compound 9a | | Descriptor: | 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-[(phenylmethyl)amino]hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
6TSM
 
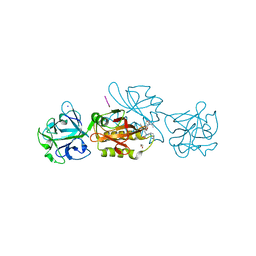 | | Marasmius oreades agglutinin (MOA) in complex with the truncated PVVRAHS synthetic substrate | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
6E61
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6TSO
 
 | | Marasmius oreades agglutinin (MOA) inhibited by cadmium | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CADMIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
