7OB3
 
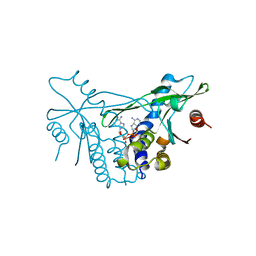 | | hSTING in complex with 3',3'-c-di-araAMP | | Descriptor: | 3',3'-c-di-araAMP, Stimulator of interferon genes protein | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2021-04-20 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Synthesis of 3'-5', 3'-5' Cyclic Dinucleotides, Their Binding Properties to the Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations
Biochemistry, 2021
|
|
3FAT
 
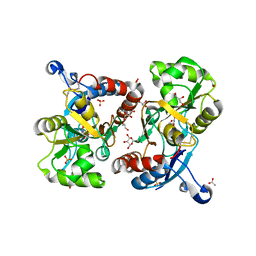 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-AMPA at 1.90A resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
5V2P
 
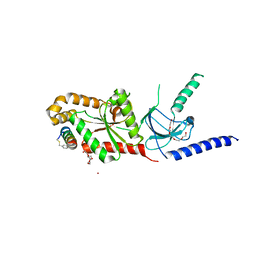 | | CaV beta2a subunit: CaV1.2 AID-CAP complex | | Descriptor: | 1,3-bis(bromomethyl)benzene, NICKEL (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Findeisen, F, Campiglio, M, Jo, H, Rumpf, C.H, Pope, L, Flucher, B, Degrado, W.F, Minor, D.L. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stapled Voltage-Gated Calcium Channel (CaV) alpha-Interaction Domain (AID) Peptides Act As Selective Protein-Protein Interaction Inhibitors of CaV Function.
ACS Chem Neurosci, 8, 2017
|
|
7AE2
 
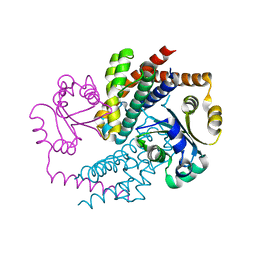 | | Crystal structure of HEPN(H107A-Y109F) toxin in complex with MNT antitoxin | | Descriptor: | HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | Descriptor: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6U
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | Descriptor: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
5VI5
 
 | | Structure of Mycobacterium smegmatis transcription initiation complex with a full transcription bubble | | Descriptor: | 1,2-ETHANEDIOL, DNA (44-MER), DNA (49-MER), ... | | Authors: | Darst, S.A, Campbell, E.A, Lilic, M. | | Deposit date: | 2017-04-14 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures.
Nat Commun, 8, 2017
|
|
6T82
 
 | |
5W6B
 
 | | Phosphotriesterase variant S1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-06-16 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Phosphotriesterase variant S1
To Be Published
|
|
7B0C
 
 | | [4Fe-4S]-NsrR complexed to 23-bp HmpA1 operator fragment | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*GP*AP*AP*TP*AP*TP*CP*AP*TP*CP*TP*AP*CP*CP*AP*AP*TP*T)-3'), DNA (5'-D(P*AP*AP*TP*TP*GP*GP*TP*AP*GP*AP*TP*GP*AP*TP*AP*TP*TP*CP*GP*TP*GP*TP*T)-3'), HTH-type transcriptional repressor NsrR, ... | | Authors: | Rohac, R, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2020-11-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors.
Commun Biol, 5, 2022
|
|
2QY1
 
 | | pectate lyase A31G/R236F from Xanthomonas campestris | | Descriptor: | PHOSPHATE ION, Pectate lyase II | | Authors: | Garron, M.L, Shaya, D. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of the thermostability and activity of a pectate lyase by single amino acid substitutions, using a strategy based on melting-temperature-guided sequence alignment.
Appl.Environ.Microbiol., 74, 2008
|
|
7U57
 
 | | apo-CTX-M-15 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-03-01 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U49
 
 | | DFC-CTX-M-15 | | Descriptor: | (2R,4S,5R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-oxoethyl]-5-(sulfanylmethyl)-1,3-thiazinane-4-carboxylic acid, Beta-lactamase | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U48
 
 | | Clavulanic acid-CTX-M-15 | | Descriptor: | (E)-5-hydroxy-3-oxo-N-(3-oxopropylidene)-L-norvaline, Beta-lactamase, GLYCEROL, ... | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
5WCW
 
 | | Phosphotriesterase variant S3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-02 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | Phosphotriesterase variant S4
To Be Published
|
|
5WFY
 
 | |
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
5WCQ
 
 | | Phosphotriesterase variant S2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Phosphotriesterase variant S2
To Be Published
|
|
5WE1
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WJ0
 
 | | Phosphotriesterase variant S5+254R | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase variant S5+254R
To Be Published
|
|
8ITE
 
 | |
5WCP
 
 | | Phosphotriesterase variant S7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphotriesterase variant S7
To Be Published
|
|
5WMS
 
 | | Phosphotriesterase variant S7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphotriesterase variant S7
To Be Published
|
|
4HI4
 
 | |
