6RD7
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 1, focussed refinement of Fo and peripheral stalk | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6ZHW
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 1 ps structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Branden, G, Neutze, R. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
6GGG
 
 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | 2-[(3~{S})-7-fluoranyl-6-(2-methylpropyl)-4-[(3-oxidanylidene-4~{H}-1,4-benzoxazin-6-yl)carbonyl]-2,3-dihydro-1,4-benzoxazin-3-yl]-~{N}-methyl-ethanamide, CHLORIDE ION, Mineralocorticoid receptor, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
6RDU
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REC
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9GL9
 
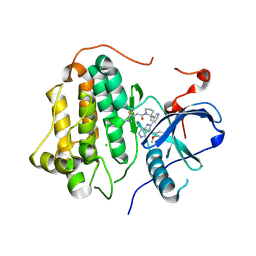 | | Wild-type EGFR bound with STX-721 | | Descriptor: | (2R,3S)-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-[(E)-4-(dimethylamino)but-2-enoyl]-2-methyl-pyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,2,3,5,6,7-hexahydropyrrolo[3,2-c]pyridin-4-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 31, 2025
|
|
8BC8
 
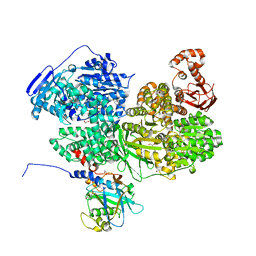 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-4-oxidanyl-benzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7NDY
 
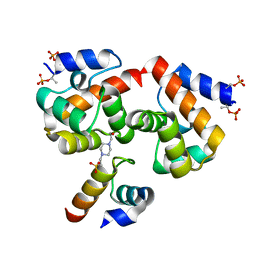 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
5A4E
 
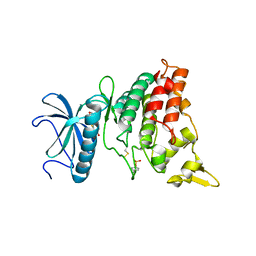 | | DYRK1A in complex with methoxy benzothiazole fragment | | Descriptor: | DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION-REGULATED KINASE 1A, N-(5-methoxy-1,3-benzothiazol-2-yl)ethanamide | | Authors: | Rothweiler, U. | | Deposit date: | 2015-06-08 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Probing the ATP-Binding Pocket of Protein Kinase Dyrk1A with Benzothiazole Fragment Molecules
J.Med.Chem., 59, 2016
|
|
6RDA
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, monomer-masked refinement | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-2: Polytomella F-ATP synthase associated subunit 2, ASA-9: Polytomella F-ATP synthase associated subunit 9, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5XKR
 
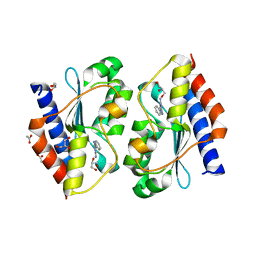 | | Crystal structure of Msmeg3575 in complex with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ACETATE ION, CMP/dCMP deaminase, ... | | Authors: | Gaded, V.M, Anand, R. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
6RE6
 
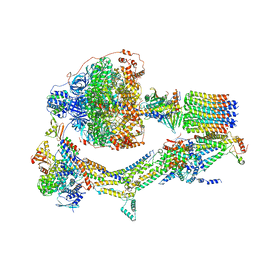 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2C, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7NNS
 
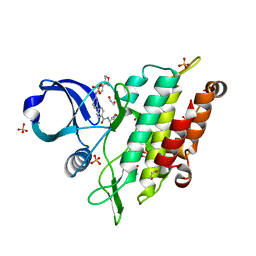 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
3FAA
 
 | |
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
5N2M
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
7RPY
 
 | | X25-2 domain of Sca5 from Ruminococcus bromii | | Descriptor: | ACETATE ION, Cohesin-containing protein, GLYCEROL, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome.
J.Biol.Chem., 298, 2022
|
|
9H91
 
 | | Cryo-EM structure of the Vibrio natrigens 50S ribosomal subunit in complex with the proline-rich antimicrobial peptide Bac5(1-17). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Raulf, K.F, Koller, T.O, Beckert, B, Morici, M, Lepak, A, Bange, G, Wilson, D.N. | | Deposit date: | 2024-10-29 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of the Vibrio natriegens 70S ribosome in complex with the proline-rich antimicrobial peptide Bac5(1-17).
Nucleic Acids Res., 53, 2025
|
|
8RNG
 
 | | Crystal structure of HLA B*18:01 in complex with TEVETYVL, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGNESIUM ION, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-10 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
4ZVF
 
 | | Crystal structure of GGDEF domain of the E. coli DosC - form II (GTP-alpha-S-bound) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Diguanylate cyclase DosC, ... | | Authors: | Tarnawski, M, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural analysis of an oxygen-regulated diguanylate cyclase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8A1E
 
 | | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | | Descriptor: | Fab 1112-1 heavy chain variable domain, Fab 1112-1 light chain variable domain, Fab 17C7 heavy chain variable domain, ... | | Authors: | Ng, W.M, Fedosyuk, S, English, S, Augusto, G, Berg, A, Thorley, L, Haselon, A.S, Segireddy, R.R, Bowden, T.A, Douglas, A.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Cell Host Microbe, 30, 2022
|
|
6P1A
 
 | |
7ZWX
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-[1,3-benzodioxol-5-ylmethyl(methyl)amino]-1-~{tert}-butyl-5~{H}-pyrazolo[3,4-d]pyrimidin-4-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
4ZS8
 
 | | Crystal structure of ligand-free, full length DasR | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional repressor DasR | | Authors: | Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Global Regulator DasR from Streptomyces coelicolor: Implications for the Allosteric Regulation of GntR/HutC Repressors.
Plos One, 11, 2016
|
|
