8AHI
 
 | |
8ALP
 
 | | Botulinum neurotoxin A6 cell binding domain crystal form II | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-08-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
9F6Z
 
 | |
5J9Z
 
 | | EGFR-T790M in complex with pyrazolopyrimidine inhibitor 1a | | Descriptor: | (R)-1-(3-(4-amino-3-(1-methyl-1H-indol-3-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Becker, C, Engel, J, Rauh, D. | | Deposit date: | 2016-04-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the Inhibition of Drug-Resistant Mutants of the Receptor Tyrosine Kinase EGFR.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
9GQI
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(11,6)-(E)-OH | | Descriptor: | (2~{S},9~{S},12~{R},13~{S},14~{E},18~{R})-2-cyclohexyl-22,23-dimethoxy-12,18-dimethyl-13-oxidanyl-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5ZKQ
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7ROO
 
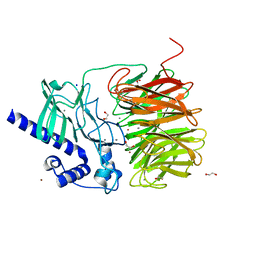 | |
6TID
 
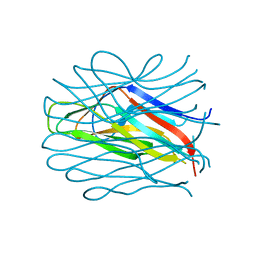 | |
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9GPS
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/p | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-19-methyl-11,20-dioxa-5-aza-1lambda5-polona-1,19lambda5-dithia-18-azanida-1lambda5-stannapentacyclo[19.2.1.05,10.013,18.018,22]tetracosane-4,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPZ
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/o | | Descriptor: | 3-cyclohexyl-17,18-dimethoxy-13-methyl-17-methylol-15,19-dioxa-1,17lambda5-diphospha-1,11lambda5-distannapentacyclo[15.3.1.04,9.011,13.013,19]heneicosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FF7
 
 | | Structure of the BMOE-crosslinked transcription termination factor Rho in the presence of ppGpp; S84C/M405C double mutant | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2024-05-22 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nucleotide-induced hyper-oligomerization inactivates transcription termination factor rho.
Nat Commun, 16, 2025
|
|
9GQC
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(6,4)-(E) | | Descriptor: | (2~{S},9~{S},12~{R},14~{E})-2-cyclohexyl-23,24-dimethoxy-12-methyl-11,17,21-trioxa-4-azatricyclo[20.3.1.0^{4,9}]hexacosa-1(26),14,22,24-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPQ
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12h/p | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-17-methyl-11,18-dioxa-1,17lambda5-diphospha-19lambda5-stannapentacyclo[17.3.1.04,9.04,13.015,20]tricosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7ZW8
 
 | |
8JT6
 
 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
9GPO
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/n | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-17-methyl-11,15-dioxa-1-aza-1lambda5-polona-1,18-distannapentacyclo[15.7.0.01,10.010,10.011,16]eicosane-2,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8EJV
 
 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and structural characterization of an IclR family transcription factor for the development of dicarboxylic acid biosensors.
Febs J., 291, 2024
|
|
9GQF
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(2,3)-(E) | | Descriptor: | (2~{S},9~{S},14~{E})-2-cyclohexyl-22,23-dimethoxy-11,17,20-trioxa-4-azatricyclo[19.3.1.0^{4,9}]pentacosa-1(25),14,21,23-tetraene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Spiske, M, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally Restricted Macrocycles as Improved FKBP51 Inhibitors Enabled by Systematic Linker Derivatization.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9F8M
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl][4-(propylamino)phenyl]methanone | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phenazine biosynthesis protein A/B, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9EW1
 
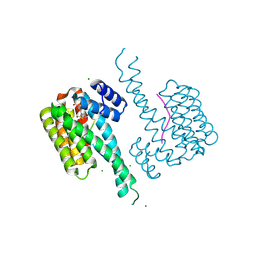 | | Ternary structure of 14-3-3s, CRAF phosphopeptide (pS259) and compound 79 (1124379). | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-1-[8-(4-iodophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]ethanone, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
7ZO2
 
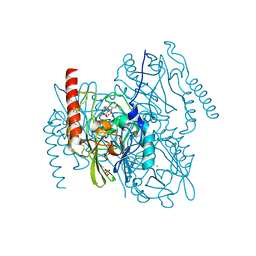 | | L1 metallo-beta-lactamase complex with hydrolysed doripenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase L1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
9NN9
 
 | | ISG15 complexed with nanobody | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION, ... | | Authors: | Dabhade, P, Schwartz, T.U. | | Deposit date: | 2025-03-05 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Identification and characterization of nanobodies specific for the human ubiquitin-like ISG15 protein.
J.Biol.Chem., 301, 2025
|
|
