1V90
 
 | | Solution structure by NMR means of delta-paluIT1-NH2 | | Descriptor: | Delta-palutoxin IT1 | | Authors: | Ferrat, G, Bosmans, F, Tytgat, J, Pimentel, C, Chagot, B, Nakajima, T, Darbon, H, Corzo, G. | | Deposit date: | 2004-01-19 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two insect-specific spider toxins and their pharmacological interaction with the insect voltage-gated Na(+) channel
Proteins, 59, 2005
|
|
2MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
1KDU
 
 | | SEQUENTIAL 1H NMR ASSIGNMENTS AND SECONDARY STRUCTURE OF THE KRINGLE DOMAIN FROM UROKINASE | | Descriptor: | PLASMINOGEN ACTIVATOR | | Authors: | Li, X, Bokman, A.M, Llinas, M, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kringle domain from urokinase-type plasminogen activator.
J.Mol.Biol., 235, 1994
|
|
1LV1
 
 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1YKG
 
 | | Solution structure of the flavodoxin-like domain from the Escherichia coli sulfite reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sulfite reductase [NADPH] flavoprotein alpha-component | | Authors: | Sibille, N, Blackledge, M, Brutscher, B, Coves, J, Bersch, B. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Sulfite Reductase Flavodoxin-like Domain from Escherichia coli
Biochemistry, 44, 2005
|
|
2BTX
 
 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ALPHA-BUNGAROTOXIN, LIBRARY DERIVED PEPTIDE | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2LCJ
 
 | |
1ZN5
 
 | |
2APN
 
 | |
2NEF
 
 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
2E4I
 
 | | Human Telomeric DNA mixed-parallel/antiparallel quadruplex under Physiological Ionic Conditions Stabilized by Proper Incorporation of 8-Bromoguanosines | | Descriptor: | DNA (5'-D(*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*(BGM)P*DGP*DTP*DTP*DAP*(BGM)P*DGP*DG)-3') | | Authors: | Matsugami, A, Xu, Y, Noguchi, Y, Sugiyama, H, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution
Febs J., 274, 2007
|
|
2JQD
 
 | |
2K3F
 
 | |
2ADR
 
 | |
2MI7
 
 | | Solution NMR structure of alpha3Y | | Descriptor: | de novo protein a3Y | | Authors: | Glover, S.D, Jorge, C, Liang, L, Valentine, K.G, Hammarstrom, L, Tommos, C. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photochemical tyrosine oxidation in the structurally well-defined alpha 3Y protein: proton-coupled electron transfer and a long-lived tyrosine radical.
J.Am.Chem.Soc., 136, 2014
|
|
2JZT
 
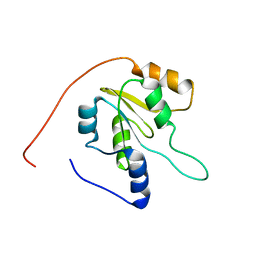 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
1ZAC
 
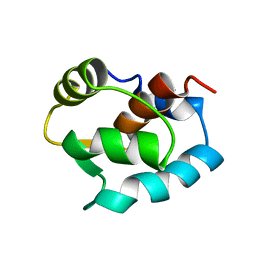 | | N-DOMAIN OF TROPONIN C FROM CHICKEN SKELETAL MUSCLE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
2LG1
 
 | |
2M5B
 
 | | The NMR structure of the BID-BAK complex | | Descriptor: | Bcl-2 homologous antagonist/killer, human_BID_BH3_SAHB | | Authors: | Moldoveanu, T, Grace, C.R, Kriwacki, R.W, Green, D.R. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | BID-induced structural changes in BAK promote apoptosis.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MCW
 
 | | Solid-state NMR structure of piscidin 3 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2021-08-18 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2GVB
 
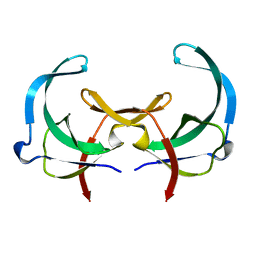 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
