252D
 
 | |
1PDO
 
 | |
1CRK
 
 | | MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Schnyder, T, Wallimann, T, Kabsch, W. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mitochondrial creatine kinase.
Nature, 381, 1996
|
|
1FBT
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
1HTT
 
 | | HISTIDYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, HISTIDYL-TRNA SYNTHETASE | | Authors: | Arnez, J.G, Harris, D.C, Mitschler, A, Rees, B, Francklyn, C.S, Moras, D. | | Deposit date: | 1996-03-09 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate.
EMBO J., 14, 1995
|
|
4AAH
 
 | | METHANOL DEHYDROGENASE FROM METHYLOPHILUS W3A1 | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE, PYRROLOQUINOLINE QUINONE | | Authors: | Mathews, F.S, Xia, Z.-X. | | Deposit date: | 1996-03-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1.
J.Mol.Biol., 259, 1996
|
|
1JAQ
 
 | | COMPLEX OF 1-HYDROXYLAMINE-2-ISOBUTYLMALONYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
1JAO
 
 | | COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2S)-2-benzyl-3-sulfanylpropanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
1JAN
 
 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (PHE79 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (PHE79 FORM), PRO-LEU-GLY-HYDROXYLAMINE INHIBITOR, ... | | Authors: | Reinemer, P, Grams, F, Huber, R, Kleine, T, Schnierer, S, Pieper, M, Tschesche, H, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural implications for the role of the N terminus in the 'superactivation' of collagenases. A crystallographic study.
FEBS Lett., 338, 1994
|
|
1JAP
 
 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), PRO-LEU-GLY-HYDROXYLAMINE, ... | | Authors: | Bode, W, Reinemer, P, Huber, R, Kleine, T, Schnierer, S, Tschesche, H. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity.
EMBO J., 13, 1994
|
|
1MRA
 
 | |
1NOJ
 
 | |
1NOK
 
 | |
1NOI
 
 | |
1GUR
 
 | | GURMARIN, A SWEET TASTE-SUPPRESSING POLYPEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | GURMARIN | | Authors: | Arai, K, Ishima, R, Morikawa, S, Imoto, T, Yoshimura, S, Aimoto, S, Akasaka, K. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of gurmarin, a sweet taste-suppressing polypeptide.
J.Biomol.NMR, 5, 1995
|
|
1HGX
 
 | |
1IDO
 
 | | I-DOMAIN FROM INTEGRIN CR3, MG2+ BOUND | | Descriptor: | INTEGRIN, MAGNESIUM ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18).
Cell(Cambridge,Mass.), 80, 1995
|
|
1ELO
 
 | | ELONGATION FACTOR G WITHOUT NUCLEOTIDE | | Descriptor: | ELONGATION FACTOR G | | Authors: | Aevarsson, A, Brazhnikov, E, Garber, M, Zheltonosova, J, Chirgadze, Yu, Al-Karadaghi, S, Svensson, L.A, Liljas, A. | | Deposit date: | 1996-03-13 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus.
EMBO J., 13, 1994
|
|
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1NIN
 
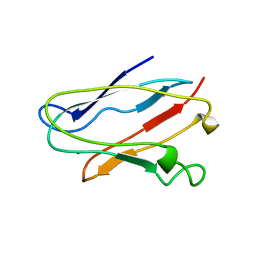 | | PLASTOCYANIN FROM ANABAENA VARIABILIS, NMR, 20 STRUCTURES | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Badsberg, U, Jorgensen, A.M.M, Gesmar, H, Led, J.J, Hammerstad-Petersen, J.M, Ulstrup, J. | | Deposit date: | 1996-03-13 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced plastocyanin from the blue-green alga Anabaena variabilis.
Biochemistry, 35, 1996
|
|
1HAB
 
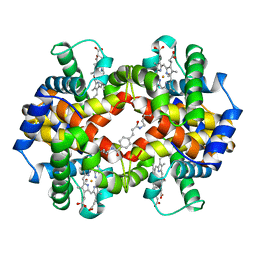 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1HAC
 
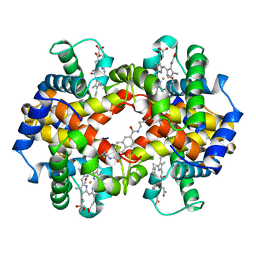 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1IOB
 
 | |
1DIN
 
 | |
1ANV
 
 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
