6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
3AKG
 
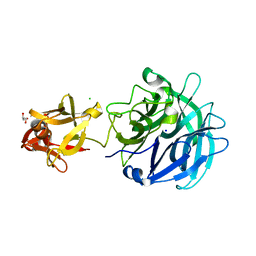 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKH
 
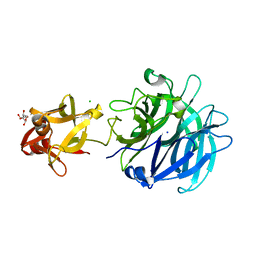 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
5T0N
 
 | |
3AKF
 
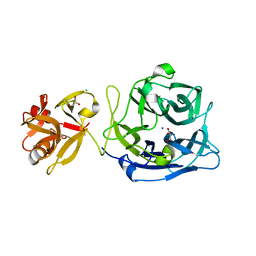 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKI
 
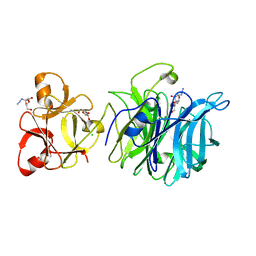 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
4TYO
 
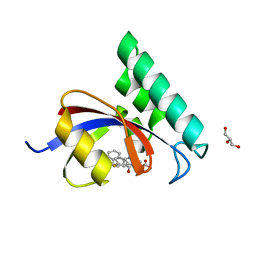 | | PPIase in complex with a non-phosphate small molecule inhibitor. | | Descriptor: | 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-D-alanine, GLYCEROL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2XHI
 
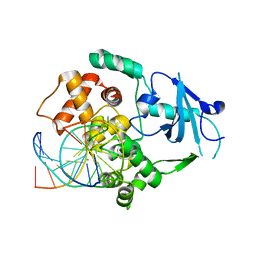 | | Separation-of-function mutants unravel the dual reaction mode of human 8-oxoguanine DNA glycosylase | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Dalhus, B, Forsbring, M, Helle, I.H, Backe, P.H, Forstrom, R.J, Alseth, I, Bjoras, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Separation-of-Function Mutants Unravel the Dual- Reaction Mode of Human 8-Oxoguanine DNA Glycosylase.
Structure, 19, 2011
|
|
8SPP
 
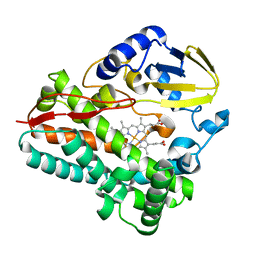 | |
2Y5Z
 
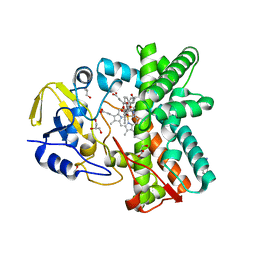 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
6EJN
 
 | |
4U85
 
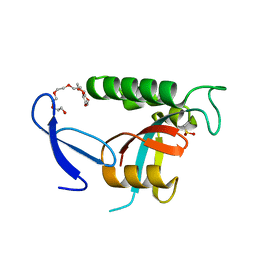 | | Human Pin1 with cysteine sulfinic acid 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U86
 
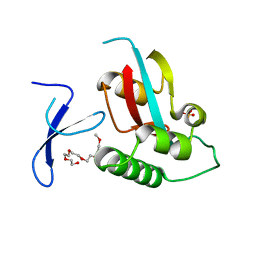 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4TVS
 
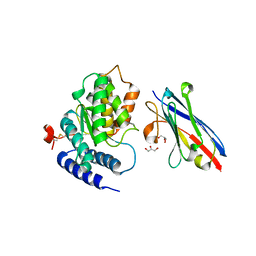 | | LAP1(aa356-583), H.sapiens, bound to VHH-BS1 | | Descriptor: | GLYCEROL, SULFATE ION, Torsin-1A-interacting protein 1, ... | | Authors: | Sosa, B.A, Schwartz, T.U. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Elife, 3, 2014
|
|
2Y46
 
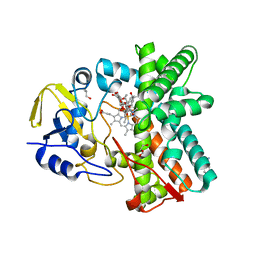 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in C 2 2 21 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y5N
 
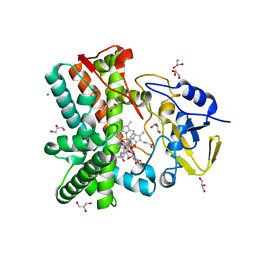 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4U84
 
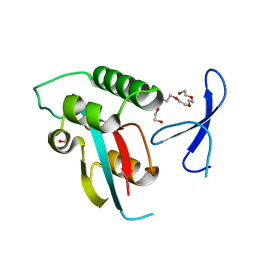 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
6F5E
 
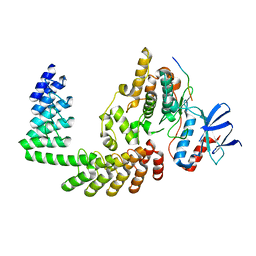 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, DD_D12_10_47, Mitogen-activated protein kinase 8 | | Authors: | Wu, Y, Mittl, P.R, Honegger, A, Batyuk, A, Plueckthun, A. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
To be published
|
|
6GFJ
 
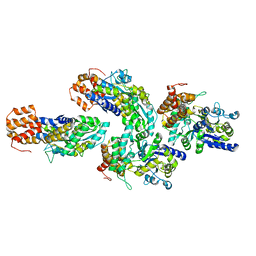 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
2KA2
 
 | |
6FUZ
 
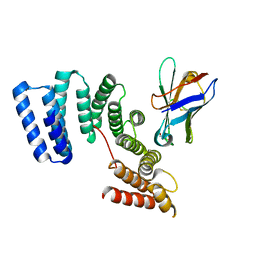 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of JIP1 | | Descriptor: | GLYCEROL, Kinesin light chain 1,Kinesin light chain 1,C-Jun-amino-terminal kinase-interacting protein 1, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
2KA1
 
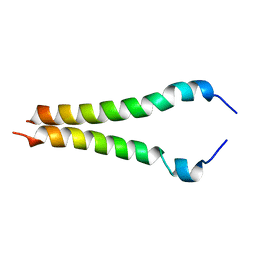 | |
6DCX
 
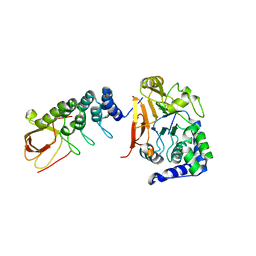 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
6YLY
 
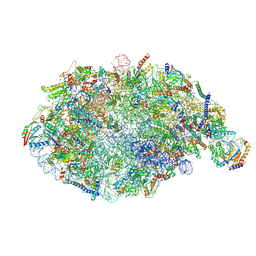 | | pre-60S State NE2 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
