8TBY
 
 | | Apo Bcs1, unsymmetrized | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TI0
 
 | | ATP-1 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T7U
 
 | | ADP-bound Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TP1
 
 | | ATP-2 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
6JQ0
 
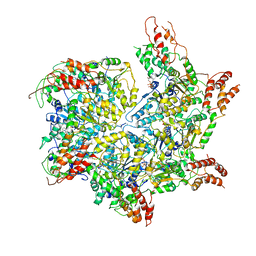 | | CryoEM structure of Abo1 Walker B (E372Q) mutant hexamer - ATP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized AAA domain-containing protein C31G5.19, ... | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
7QO6
 
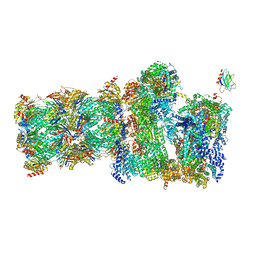 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO5
 
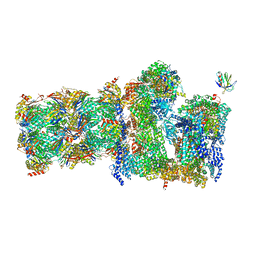 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO4
 
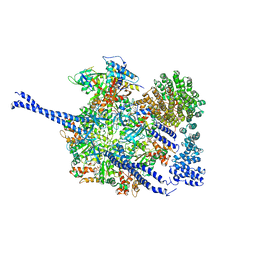 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UK9
 
 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UH7
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6KWW
 
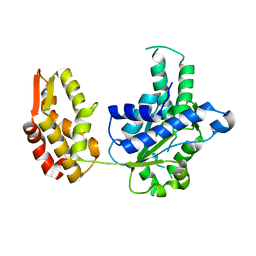 | | HslU from Staphylococcus aureus | | Descriptor: | ATP-dependent protease ATPase subunit HslU | | Authors: | Ha, N.-C, Jeong, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleavage-Dependent Activation of ATP-Dependent Protease HslUV from Staphylococcus aureus .
Mol.Cells, 43, 2020
|
|
7QXN
 
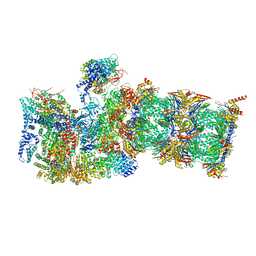 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXX
 
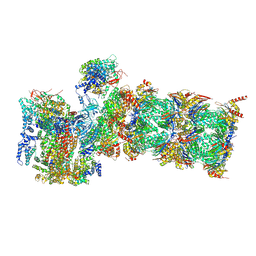 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYB
 
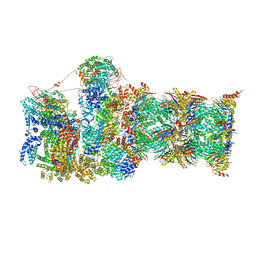 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QY7
 
 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXP
 
 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXU
 
 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXW
 
 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYA
 
 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7R7S
 
 | | p47-bound p97-R155H mutant with ATPgammaS | | Descriptor: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
8UII
 
 | |
