5NPP
 
 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3MAX
 
 | |
4MAX
 
 | | Crystal structure of Synechococcus sp. PCC 7002 globin at cryogenic temperature with heme modification | | Descriptor: | HEME B/C, SULFATE ION, cyanoglobin | | Authors: | Wenke, B.B, Schlessman, J.L, Heroux, A, Lecomte, J.T.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | The 2/2 hemoglobin from the cyanobacterium Synechococcus sp. PCC 7002 with covalently attached heme: Comparison of X-ray and NMR structures.
Proteins, 82, 2014
|
|
6MAX
 
 | |
1MAX
 
 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | Authors: | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | Deposit date: | 1996-02-06 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
2MAX
 
 | |
7MAX
 
 | |
4Y08
 
 | | ONE MINUTE IRON LOADED HUMAN H FERRITIN | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin heavy chain, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Iron binding to human heavy-chain ferritin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZJK
 
 | | FIVE MINUTES IRON LOADED HUMAN H FERRITIN | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin heavy chain, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iron binding to human heavy-chain ferritin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YKH
 
 | | Thirty minutes iron loaded human H ferritin | | Descriptor: | BICINE, CHLORIDE ION, FE (II) ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2015-03-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Iron binding to human heavy-chain ferritin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OYN
 
 | | Fifteen minutes iron loaded human H ferritin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, CHLORIDE ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2014-02-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Iron binding to human heavy-chain ferritin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1OEY
 
 | | Heterodimer of p40phox and p67phox PB1 domains from human NADPH oxidase | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 2, NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Wilson, M.I, Gill, D.J, Perisic, O, Quinn, M.T, Williams, R.L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pb1 Domain-Mediated Heterodimerization in Nadph Oxidase and Signaling Complexes of Atypical Protein Kinase C with Par6 and P62
Mol.Cell, 12, 2003
|
|
7TIK
 
 | |
1IK3
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID | | Descriptor: | (TRANS-12,13-EPOXY)-11-HYDROXY-9(Z)-OCTADECENOIC ACID, (TRANS-12,13-EPOXY)-9-HYDROXY-10(E)-OCTADECENOIC ACID, 13(R)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID, ... | | Authors: | Skrzypczak-Jankun, E, Funk Jr, M.O. | | Deposit date: | 2001-05-01 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a purple lipoxygenase.
J.Am.Chem.Soc., 123, 2001
|
|
1IPK
 
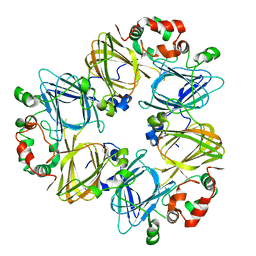 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1S08
 
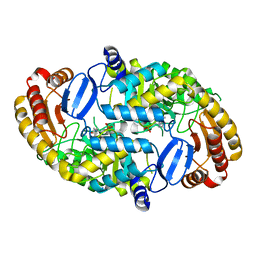 | | Crystal Structure of the D147N Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S0A
 
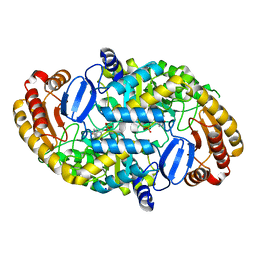 | | Crystal Structure of the Y17F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S09
 
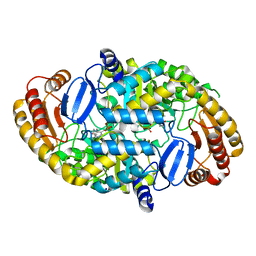 | | Crystal Structure of the Y144F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1D6R
 
 | | CRYSTAL STRUCTURE OF CANCER CHEMOPREVENTIVE BOWMAN-BIRK INHIBITOR IN TERNARY COMPLEX WITH BOVINE TRYPSIN AT 2.3 A RESOLUTION. STRUCTURAL BASIS OF JANUS-FACED SERINE PROTEASE INHIBITOR SPECIFICITY | | Descriptor: | BOWMAN-BIRK PROTEINASE INHIBITOR PRECURSOR, TRYPSINOGEN | | Authors: | Koepke, J, Ermler, U, Wenzl, G, Flecker, P. | | Deposit date: | 1999-10-15 | | Release date: | 2000-05-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
J.Mol.Biol., 298, 2000
|
|
1S6J
 
 | |
1S6I
 
 | |
8RP8
 
 | | Structure of K2 Fab in complex with human CD47 ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Laursen, M, Kelpsas, V, Rose, N. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
Mabs, 16, 2024
|
|
8RPB
 
 | | Structure of S79 Fab in complex with IgV domain of human PD-L1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Programmed cell death 1 ligand 1, ... | | Authors: | Svensson, A, Kelpsas, V, Laursen, M, Rose, N. | | Deposit date: | 2024-01-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
Mabs, 16, 2024
|
|
6Q76
 
 | |
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | Descriptor: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poulos, T.L, Kim, J, Murarka, V.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.40000689 Å) | | Cite: | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
