8FMW
 
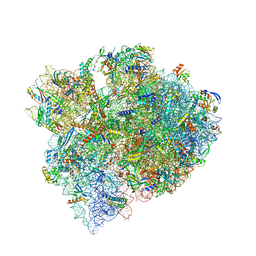 | | The structure of a hibernating ribosome in the Lyme disease pathogen | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Manjari, S.R, Agrawal, E.K, Keshavan, P, Koripella, R.K, Majumdar, S, Marcinkiewicz, A.L, Lin, Y.P, Agrawal, R.K, Banavali, N.K. | | Deposit date: | 2022-12-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The structure of a hibernating ribosome in a Lyme disease pathogen.
Nat Commun, 14, 2023
|
|
8FMJ
 
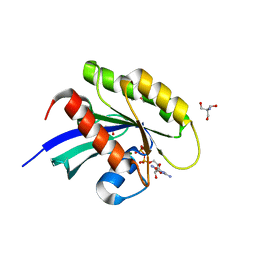 | | Crystal structure of human KRAS in space group R32 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Brenner, R, Landgraf, A, Gonzalez-Gutierrez, G, Bum-Erdene, K, Meroueh, S.O. | | Deposit date: | 2022-12-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal Packing Reveals a Potential Autoinhibited KRAS Dimer Interface and a Strategy for Small-Molecule Inhibition of RAS Signaling.
Biochemistry, 62, 2023
|
|
8FMH
 
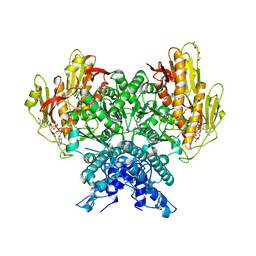 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMF
 
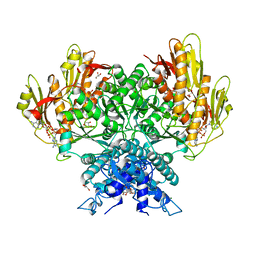 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FLW
 
 | | Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8FLO
 
 | | X-ray crystal structure of substrate free CYP124A1 from Mycobacterium Marinum | | Descriptor: | Cytochrome P450 124A1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The catalytic activity and structure of the lipid metabolizing CYP124 cytochrome P450 enzyme from Mycobacterium marinum.
Arch.Biochem.Biophys., 737, 2023
|
|
8FLN
 
 | | Crystal structure of BTK C481S kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8FLL
 
 | | Crystal structure of BTK kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8FLH
 
 | | Bruton's tyrosine kinase in complex with an orthosteric inhibitor | | Descriptor: | 2-(3,5-dichloroanilino)-1-{(3R)-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]azepan-1-yl}ethan-1-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8FL5
 
 | |
8FL1
 
 | | Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8FKS
 
 | |
8FKR
 
 | |
8FKQ
 
 | |
8FKP
 
 | |
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKG
 
 | |
8FKF
 
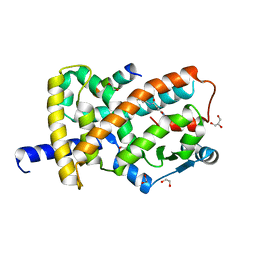 | |
8FK8
 
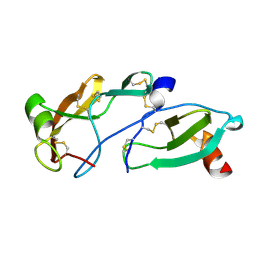 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
8FK6
 
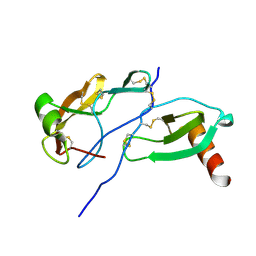 | | Crystal Structure of the Tick Evasin EVA-AAM1001(Y44A) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
8FK5
 
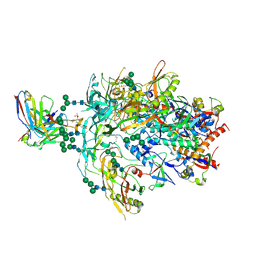 | | Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8FK3
 
 | |
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
