6FCK
 
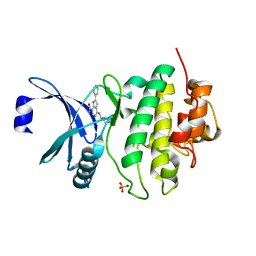 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7UMV
 
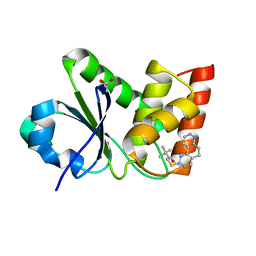 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((5,6-dihydropyrido[2,3-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 1-{[(10aP)-5,6-dihydropyrido[2,3-h]quinazolin-2-yl]sulfanyl}-3,3-dimethylbutan-2-one, ACETATE ION, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
9MSA
 
 | | Alpha-ketoisovalerate decarboxylase (Kivd) from Synechocystis sp. PCC 6803 with substitution S286T | | Descriptor: | 1,2-ETHANEDIOL, Alpha-ketoisovalerate decarboxylase, MAGNESIUM ION, ... | | Authors: | Begum, A, Xie, H, Gunn, L.H. | | Deposit date: | 2025-01-09 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Directed evolution of alpha-ketoisovalerate decarboxylase for improved isobutanol and 3-methyl-1-butanol production in cyanobacteria.
Biotechnol Biofuels Bioprod, 18, 2025
|
|
5NVW
 
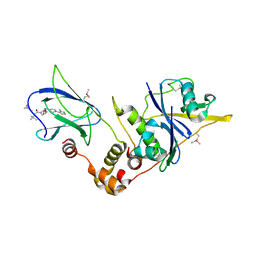 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
8BSL
 
 | | Human GLS in complex with compound 12 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, ~{N}-[5-[[(3~{R})-1-(5-azanyl-1,3,4-thiadiazol-2-yl)pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]-2-phenyl-ethanamide | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
7UN4
 
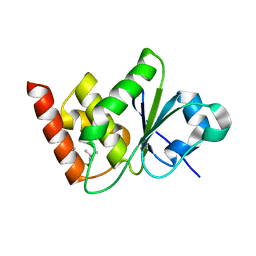 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-propyl-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 1-{[(9aM)-5,6-dihydrothieno[2,3-h]quinazolin-2-yl]sulfanyl}-3,3-dimethylbutan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
8R74
 
 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-(2-oxidanyl-1,3-thiazol-4-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J.Med.Chem., 67, 2024
|
|
6D52
 
 | |
7C7X
 
 | |
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6GQD
 
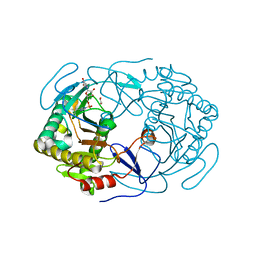 | | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Kopec, J, Bezerra, G.A, Zhang, M, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
To Be Published
|
|
6QZ7
 
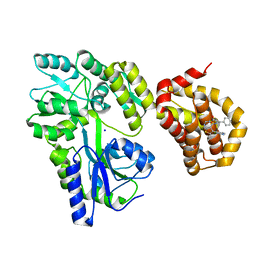 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5NW1
 
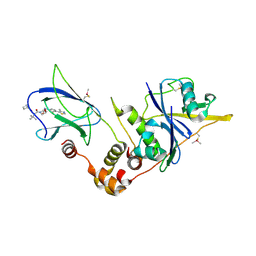 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclobutanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 18) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclobutylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
7BEA
 
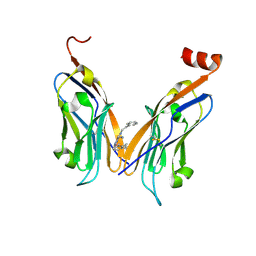 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | 2-(aminomethyl)-6-[(2-methyl-3-phenyl-phenyl)methoxy]-~{N}-(2-phenylethyl)imidazo[1,2-a]pyridin-3-amine, Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Butera, R, Wazynska, M, Holak, T, Domling, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists.
Acs Med.Chem.Lett., 12, 2021
|
|
6I1O
 
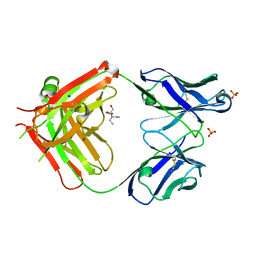 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
9LS8
 
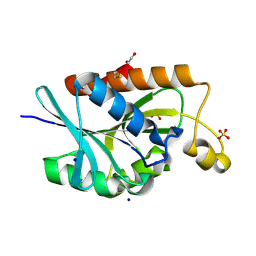 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium at 1.22 A | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Arora, A. | | Deposit date: | 2025-02-04 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int J Biol Macromol, 275, 2024
|
|
5YRS
 
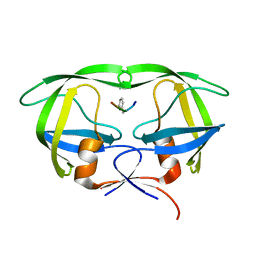 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
7YKO
 
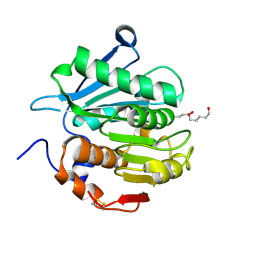 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
5J49
 
 | |
9GIZ
 
 | | UMG-SP-1, a promiscuous hydrolase | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, UMG-SP-1 | | Authors: | Tokoli, A, Stojanovski, G, Jodaitis, L, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2024-08-20 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of UMG-SP-1 at 1.89 Angstroms resolution
To Be Published
|
|
7C62
 
 | |
9MXB
 
 | | Crystal Structure of WT HIV-1 Reverse Transcriptase in Complex with 5-{2-[2-(2-oxo-4-sulfanylidene-3,4-dihydropyrimidin-1(2H)-yl)ethoxy] phenoxy}naphthalene-2-carbonitrile (JLJ648), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2-oxo-4-sulfanylidene-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, CHLORIDE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2025-01-17 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanistic basis for a novel dual-function Gag-Pol dimerizer potentiating CARD8 inflammasome activation and clearance of HIV-infected cells.
Npj Drug Discov, 2, 2025
|
|
5M99
 
 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
7ZUN
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone compound | | Descriptor: | (4~{S})-4-(2-azanylethyl)-6-phenyl-7-[3-(trifluoromethyloxy)phenyl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1-ol, Isoform 2 of Serine/threonine-protein kinase pim-1 | | Authors: | Casale, E. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Chirality, 34, 2022
|
|
7SOY
 
 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
