7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7YC0
 
 | | Acetylesterase (LgEstI) W.T. | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7Y1B
 
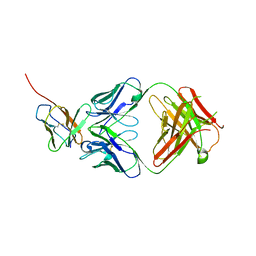 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
3CGO
 
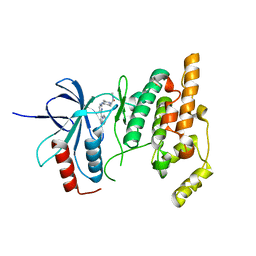 | | IRAK-4 Inhibitors (Part II)- A structure based assessment of imidazo[1,2 a]pyridine binding | | Descriptor: | 2-{4-[(4-imidazo[1,2-a]pyridin-3-ylpyrimidin-2-yl)amino]piperidin-1-yl}-N-methylacetamide, Mitogen-activated protein kinase 10 | | Authors: | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Beevers, R. | | Deposit date: | 2008-03-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | IRAK-4 inhibitors. Part II: A structure-based assessment of imidazo[1,2-a]pyridine binding
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7YVT
 
 | |
3CQH
 
 | |
3G90
 
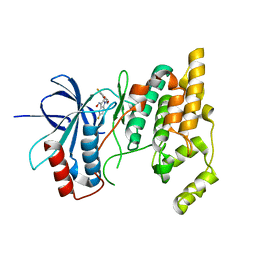 | |
3CGF
 
 | | IRAK-4 Inhibitors (Part II)- A structure based assessment of imidazo[1,2 a]pyridine binding | | Descriptor: | Mitogen-activated protein kinase 10, N-cyclohexyl-4-imidazo[1,2-a]pyridin-3-yl-N-methylpyrimidin-2-amine | | Authors: | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Buckley, G. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | IRAK-4 inhibitors. Part II: A structure-based assessment of imidazo[1,2-a]pyridine binding
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7ZKS
 
 | | SRPK1 IN COMPLEX WITH INHIBITOR | | Descriptor: | CHLORIDE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, SRSF protein kinase 1 | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
7XQT
 
 | | The structure of FLA-K*00701/KP-FECV-11 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQS
 
 | | The structure of FLA-K*00701/KP-CoV-9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide from Spike glycoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
7XQU
 
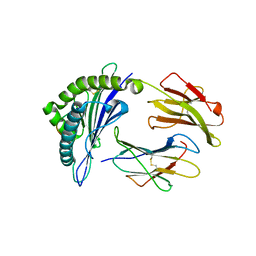 | | The structure of FLA-E*00301/EM-FECV-10 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide from Nucleoprotein | | Authors: | Qiao, P.W, Yue, C, Peng, W.Y, Liu, K.F, Huo, S.T, Zhang, D, Chai, Y, Qi, J.X, Sun, Z.Y, Gao, G.F, Liu, W.J, Wu, G.Z. | | Deposit date: | 2022-05-08 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of the characteristics of feline major histocompatibility complex class I molecules cross-presenting coronavirus peptides
To Be Published
|
|
8ASN
 
 | |
8AXZ
 
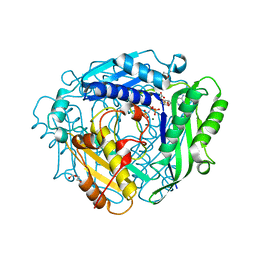 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with S-adenosylmethionine, adenosin and diphosphono-aminophosphonic acid. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L. | | Deposit date: | 2022-09-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.154 Å) | | Cite: | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with S-adenosylmethionine, adenosin and diphosphono-aminophosphonic acid.
To Be Published
|
|
8A7I
 
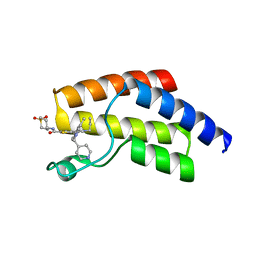 | | Crystal structure of BRD9 bromodomain in complex with compound EA-89 | | Descriptor: | Bromodomain-containing protein 9, ~{N}-[1,1-bis(oxidanylidene)thian-4-yl]-7-[3-methyl-1-(piperidin-4-ylmethyl)indol-5-yl]-4-oxidanylidene-5-propyl-thieno[3,2-c]pyridine-2-carboxamide | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BRD9 degraders as chemosensitizers in acute leukemia and multiple myeloma.
Blood Cancer J, 12, 2022
|
|
8AR7
 
 | |
8AR8
 
 | |
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8B02
 
 | | Crystal structure of the dsRBD domain of tRNA-dihydrouridine(20) synthase from Amphimedon queenslandica | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DRBM domain-containing protein, ... | | Authors: | Pecqueur, L, Faivre, B, Hamdane, D. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family.
Biomolecules, 12, 2022
|
|
8A9X
 
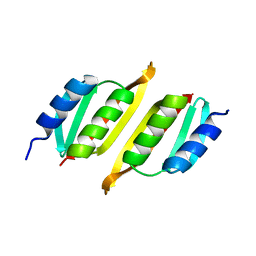 | | Crystal structure of PulM C-ter domain | | Descriptor: | Type II secretion system protein M | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
3OC9
 
 | |
3NTN
 
 | | Crystal Structure of UspA1 head and neck domain from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Conners, R, Zaccai, N, Agnew, C, Burton, N, Brady, R.L. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NZ1
 
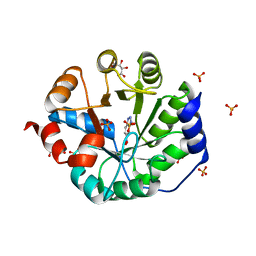 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
